Successfully Added to Cart
Customers also bought...
-
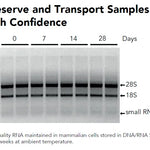 DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the...
DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the... -
 DNA/RNA Shield SafeCollect Swab Collection Kit (1 ml fill) (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA...
DNA/RNA Shield SafeCollect Swab Collection Kit (1 ml fill) (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA...
Highlights
- Robust enrichment & immunoprecipitation of DNA containing 5mC.
- Includes a highly specific anti-5-methylcytosine monoclonal antibody for defined, reproducible results.
- Eluted, ultra-pure DNA is ideal for use in subsequent molecular-based analyses (e.g., assembling genomic libraries and determining genome-wide methylation status).
Original Manufacturer
Satisfaction 100% guaranteed, read Our Promise
Innovated in California, Made in the USA
Highlights
- Robust enrichment & immunoprecipitation of DNA containing 5mC.
- Includes a highly specific anti-5-methylcytosine monoclonal antibody for defined, reproducible results.
- Eluted, ultra-pure DNA is ideal for use in subsequent molecular-based analyses (e.g., assembling genomic libraries and determining genome-wide methylation status).
Original Manufacturer
Satisfaction 100% guaranteed, read Our Promise
Innovated in California, Made in the USA
| Cat # | Name | Size | Price | |
|---|---|---|---|---|
| D5101-3-20 | MIP Buffer | 20 ml | €32,60 | |
| D5101-4-1 | DNA Denaturing Buffer | 1 ml | €20,40 | |
| A3002-50 | Anti-5-Methylcytosine Monoclonal Antibody (Clone 7D21) | 50 µl | €199,50 | |
| D5101-2 | Methylated/Non-methylated Control DNA & Primer Set | 1 Set | €148,60 | |
| D3004-4-10 | DNA Elution Buffer | 10 ml | €17,40 | |
Description
Performance
Technical Specifications
| Applicable For | Immunoprecipitation of methylated DNA, PCR, sequencing, microarrays, etc. |
|---|---|
| Elution Volume | ≥ 15 µl |
| Input | 50 - 500 ng of DNA |
| Processing Time | 4 hours |
Resources
Documents
Citations
The authors utilized the DNA Methylation IP Kit (MeDIP) from Zymo Research to compare DNA methylation results using either MeDIP-Seq or Infinium HumanMethylation450 BeadChIP array. They found the MeDIP-Seq resulted in detection of 15,709 differentially methylation regions, almost twice as many compared the array based method (8070), and concluded that MeDIP-Seq provided a more comprehensive picture of the methylation in human samples.
Clark C, Palta P, Joyce CJ, Scott C, Grundberg E, Deloukas P, Palotie A, Coffey A. 2012. A Comparison of the Whole Genome Approach of MeDIP-Seq to the Targeted Approach of the Infinium HumanMethylation450 BeadChIP for Methylome Profiling. PLOS ONE 7:11 (e5Researchers used the Methylated DNA IP Kit from Zymo Research to immunoprecipitate methylated DNA from leaf tissue and study the DNA methylation profile in the maize genome. The authors found that specific families of retrotransposons could spread heterochromatin to neighboring genes and epigenetically suppress their expression.
Eichten SR et al. (2012) Spreading of heterochromatin is limited to specific families of maize retrotransposons. PLoS Genet. 8(12):e1003127.The Methylated-DNA IP Kit was used for CpG immunoprecipitation of genomic DNA isolated from T cells that was sheared to ~200 bp using dsDNA Shearase Plus (E2018) to study the DNA methylation status of the Notch-1 promoter.
Rauen T et al. (2012) AMP-responsive element modulator α (CREMα) contributes to decreased Notch-1 expression in T cells from patients with active systemic lupus erythematosus (SLE). J Biol Chem. 287(51):42525-32.Read More
Need help? Contact Us




