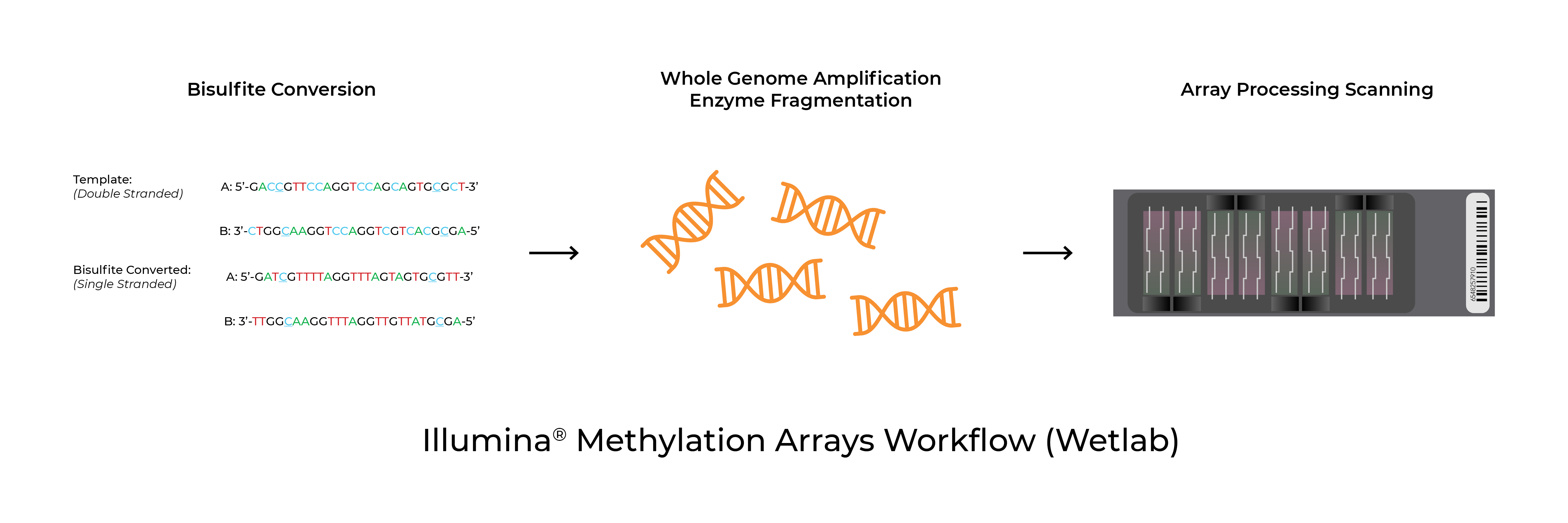
Bisulfite Conversion for Illumina Methylation Arrays
Troubleshooting & Best Practice Guidelines
Methylation arrays are common platforms for analyzing 5-methylcytosine. The Infinium™ MethylationEPIC BeadChip and Infinium™ HumanMethylation450 BeadChip (commonly referred to as the EPIC and 450K arrays respectively) as well as the recently launched Infinium™ Mouse Methylation BeadChip from Illumina® all utilize Zymo Research’s bisulfite conversion technologies to distinguish 5mC from unmodified cytosines. In addition to the test probe sets, the arrays include bisulfite conversion quality control probes. In some instances, the analysis software will flag samples for low bisulfite conversion efficiency. Possible causes for these warnings are low bisulfite conversion efficiency, low DNA input/quality, and chip failure.
Validated Protocols
The only approved and validated bisulfite conversion kits for the Infinium™ Methylation arrays are the EZ DNA Methylation Kit (catalog numbers: D5001, D5002, D5004) and the EZ DNA Methylation-Lightning Kit in magbead format only (catalog numbers: D5046, D5047, D5049). It is critical to follow the Illumina® recommended incubation protocol (16 cycles of 95°C for 30 seconds, 50°C for 60 minutes) when using the D5001, D5002, and D5004 kits. Additional details can be found in the appendix of the respective bisulfite conversion protocols. The D5046 and D5047 kits are ideal for manual preparation while the D5049 kit features optimized buffer volumes that allow for consistent high-throughput automation performance. While other bisulfite conversion kits will result in high-quality bisulfite converted DNA, only samples processed using the validated kits are supported by Illumina®.
Along with using the validated bisulfite conversion kits, it is important to follow the rest of the BeadChip protocol as written, including the downstream software analysis. Illumina® periodically reviews the performance of the analysis software and probe performance. Use the most up-to-date software and manifest files to ensure that downstream analysis is correct.
DNA Input & Quality

The amount and quality of genomic DNA used in these assays is critical to success. For best results, DNA should be as intact and high quality as possible. The arrays rely on a signal to noise ratio to determine the conversion efficiency. When low input or low quality DNA is used, the signal to noise ratio is much lower and can affect the reliability of the bisulfite conversion control signals.
The required minimum amount of DNA is 250 ng for the manual protocol and 1000 ng for the automated protocol. Genomic DNA should be quantified using a dsDNA specific method such as Picogreen® or Qubit®. NanoDrop® or other spectrophotometric methods are not recommended for quantification due to issues with successfully distinguishing DNA from RNA. RNase treatment can also help ensure that DNA quantification is accurate.
When working with DNA isolated from FFPE samples or other degraded DNA samples, 500 ng DNA inputs or higher are highly recommended. Additionally, single-column bisulfite conversion (vs. the 96-well plate) is recommended for degraded samples due to the ability to use smaller elution volumes. Bisulfite-converted FFPE DNA should then be treated with the Illumina Infinium™ FFPE DNA Restoration Kit (as described in the manual) before processing the array. The entire sample should be used.
Low Bisulfite Conversion Efficiency

Bisulfite conversion can be impacted by a variety of factors. The quality of the CT Conversion Reagent is critical for successful conversion. It is recommended to prepare the reagent fresh before each conversion if possible. Otherwise, the prepared reagent should be stored according to the guidelines listed in the protocol. The conversion reagent should not be exposed to light or oxygen any more than necessary. If processing 96-well plates, a multichannel pipette should be used and the prepared conversion reagent should be added last to the plate.
Conversion should be performed in a thermal cycler with a heated lid. Samples and conversion reagent should be mixed thoroughly (no observed mixing lines) and fully spun down before placing tubes in the thermal cycler. If the tubes are not fully spun down or the lid is not heated properly, precipitation may form around the lid of the PCR tube. After the incubation, any precipitation that is observed should be avoided when transferring the sample as it could contain unconverted DNA.
The desulphonation incubation should be stopped at 15 minutes (20 minutes is the absolute maximum). Leaving the desulphonation buffer on the column longer than the recommended time can result in additional degradation to the sample.
A bisulfite conversion quality control check is highly recommended. Bisulfite converted DNA can be assessed using a variety of methods. Bisulfite converted DNA can be quantified by qPCR or using the RNA setting on NanoDrop® (or similar spectrophotometric method). The expected DNA yield after bisulfite conversion is approximately 70-80%. This quantification step will help ensure that there is enough bisulfite converted DNA recovered for downstream processing. Bisulfite conversion efficiency can be assessed via colony Sanger Sequencing or via probe/TaqMan® assay. In rare instances, sample purity may be responsible for low bisulfite conversion efficiency. In these cases, re-extraction or further cleanup is recommended.
Chip Failure
On occasion, chip failures may occur. If multiple samples on a single chip have low bisulfite conversion efficiency as determined by the BeadArray Controls Reporter, this might indicate a chip failure rather than a bisulfite conversion issue. In these instances, re-running leftover bisulfite-converted sample on a new chip can resolve the issue. To further troubleshoot, a post-bisulfite conversion/ pre-array QC is helpful to determine whether to rerun the samples.
Conclusions
Methylation arrays such as the 450K and EPIC array are helpful tools for analyzing methylation changes in a variety of human and mouse samples. Whenever a sample is flagged for low bisulfite conversion, it is best to confirm that the following is true for your project:
- Validated and updated protocols were used
- DNA was as high-quality and intact as possible. Higher DNA input amounts were used for fragmented/low-quality samples
- Bisulfite conversion was performed according to best practices
| Bisulfite Conversion Kits Validated for Illumina®Methylation Arrays | ||
|---|---|---|
| Kit Name | Size | Catalog Number |
| EZ DNA Methylation Kit | 50 Rxns | D5001 |
| EZ DNA Methylation Kit | 200 Rxns | D5002 |
| EZ-96 DNA Methylation Kit | 2 x 96 Rxns, Deep-Well | D5004 |
| EZ-96 DNA Methylation-Lightning MagPrep | 4 x 96 Rxns | D5046 |
| EZ-96 DNA Methylation-Lightning MagPrep | 8 x 96 Rxns | D5047 |
| EZ DNA Methylation-Lightning Automation Kit* | 96 Rxns | D5049 |
*For automation scripts and support, email automation@zymoresearch.com
| DNA Methylation Analysis | |
| Bisulfite Methods | |
| Bisulfite Conversion | |
| NGS Platforms | |
| PCR Platforms | |
| Bisulfite-Free Methods | |
| Antibody Methods | |
| Enzymatic Methods | |
| Chromatin Analysis | |
| ChIP | |
| Chromatin Structure | |


