Overcoming Sample Collection Challenges In COVID-19 Testing
The COVID-19 pandemic has over 100,000+ of lives lost globally41 and is already placing significant strains on healthcare systems and economies across the globe. It is imperative that we control the spread of this respiratory infection in order to reduce mortality and disruption to livelihoods. Due to the high rate of transmission of COVID-19, one of the best ways to control the pandemic will be an active global surveillance program. However, in order to limit the spread of the virus and meet testing demands, sample collection, storage, and transportation will be a significant challenge.
Current testing methods for COVID-19 rely on the detection of viral RNA using reverse transcription quantitative polymerase chain reaction (RT-qPCR). Unfortunately, RNA integrity is very sensitive to external factors, such as ribonucleases, temperature, freeze-thaw (Figure 1), and pH. Due to the high volume of testing required, the viral RNA in commonly collected sample types, such as nasopharyngeal swabs, oropharyngeal swabs, and sputum, will need to be stable for extended periods of time. Moreover, once collected, samples need to be stabilized as they may encounter uncontrolled environments because of the limited availability of cold-chain storage and transport. It is also essential that the risk of COVID-19 exposure to couriers, medical staff, and testing clinicians is minimal, so the collected samples must be safe to handle. Furthermore, since COVID-19 testing workflows can differ significantly among diagnostic laboratories, the transportation media needs to be compatible with a variety of RNA extraction methods.
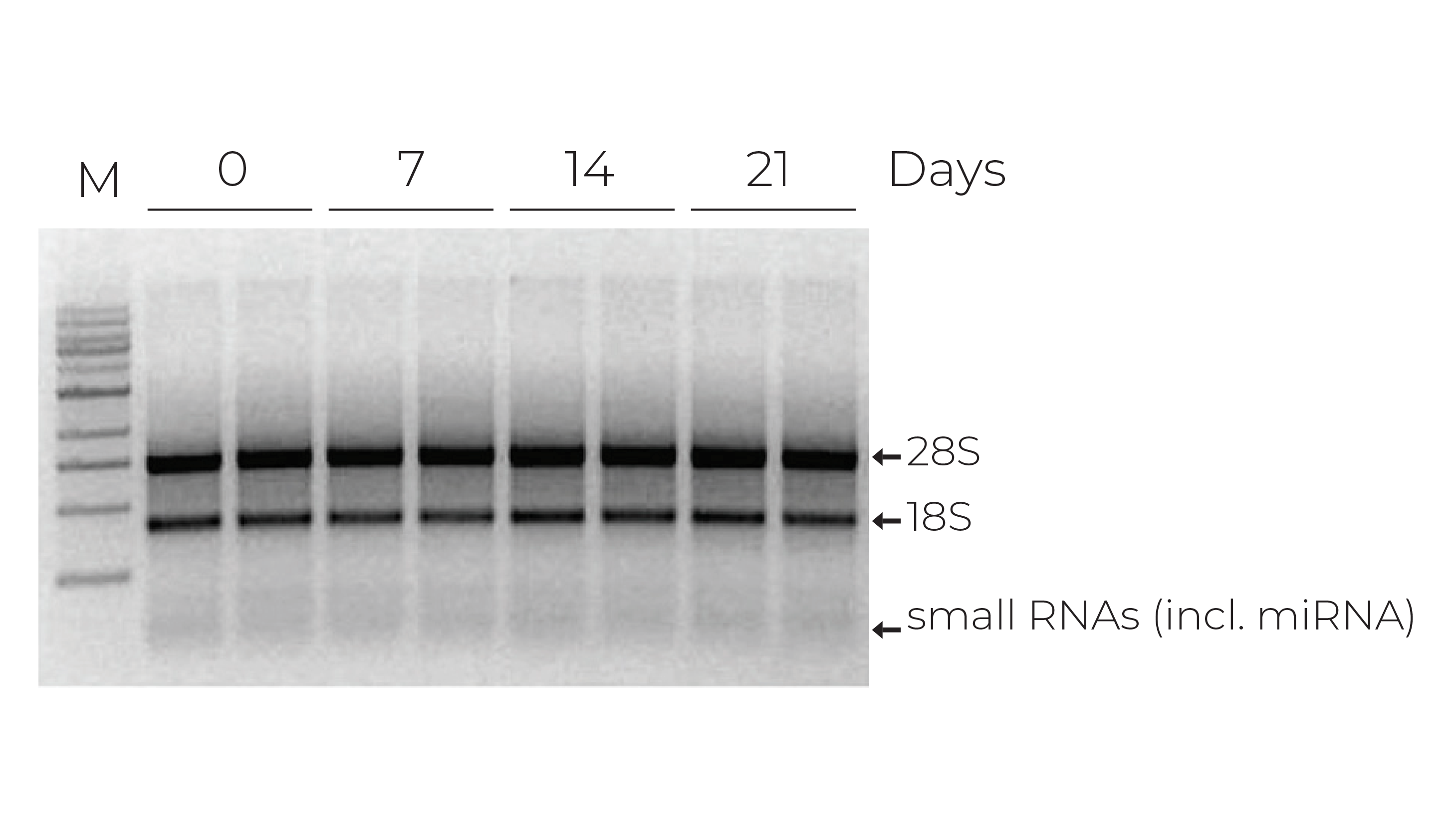
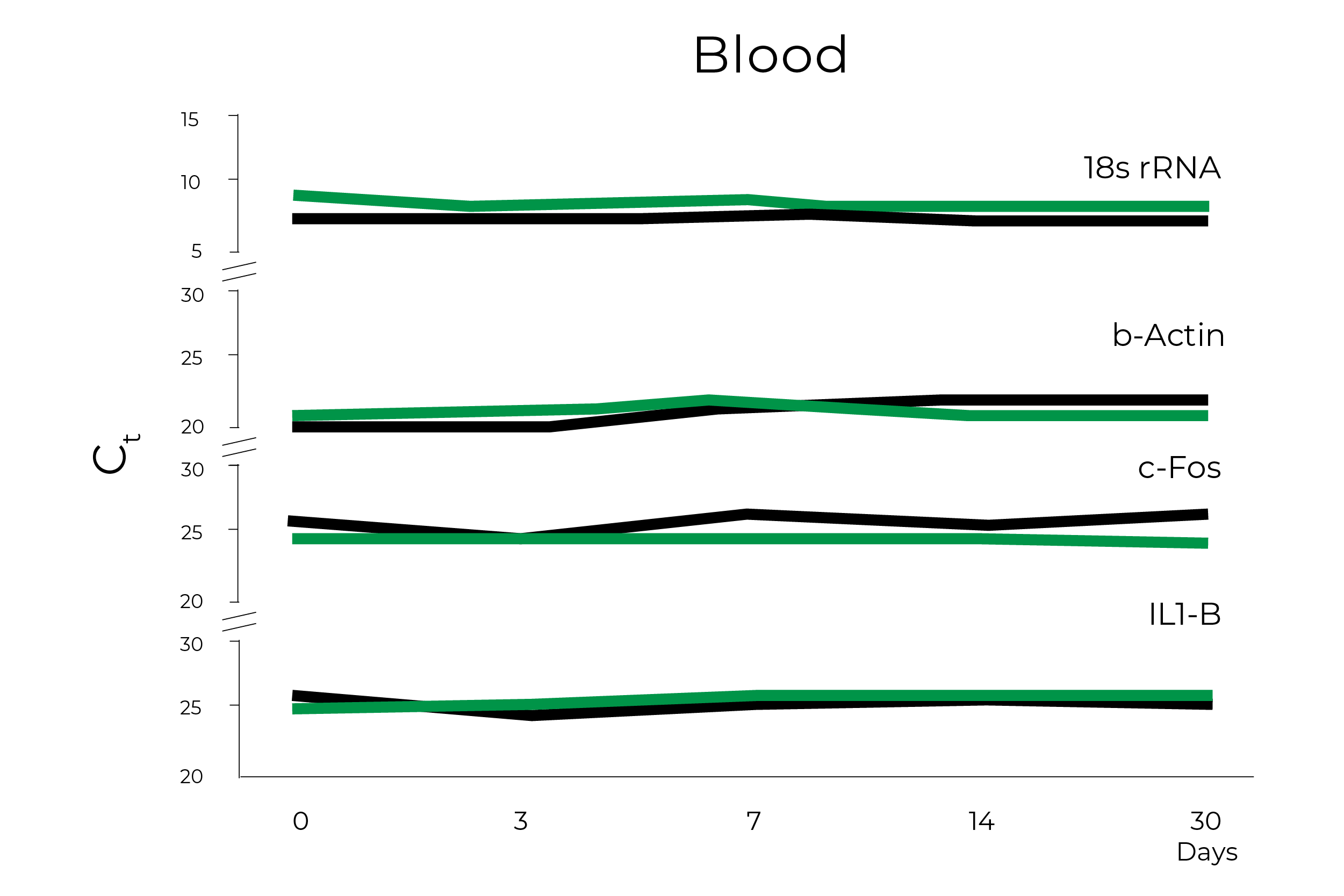
Zymo Research has been developing solutions for safely transporting and storing nucleic acid samples since 2006 and has led several efforts to support researchers and organizations with sample collection devices in this time of need. The DNA/RNA Shield sample collection stabilization solution has been an essential component in COVID-19 testing, as it can easily overcome the challenges that widespread COVID-19 testing present. Zymo Research has already clearly demonstrated that RNA within whole blood, saliva, swabs, tissues, and stool stored in DNA/RNA Shield is stable at ambient temperature for up to 30 days (Figure 2). Furthermore, an independent study comparing various pretreatment methods of whole blood prior to RNA extraction found that RNA yield, integrity, and quality were significantly better for whole blood and peripheral blood cells mixed with DNA/RNA Shield prior to storing at – 80°C1.
DNA/RNA Shield has already proven itself extensively when it comes to sample stability and assessing respiratory viral infections. DNA/RNA Shield was used in multiple studies to collect, transport, and store nasopharyngeal swabs at ambient temperatures for surveying respiratory viral infections in New York City2, 3, 4 & 5. Furthermore, the extracted RNA and DNA was suitable for viral detection via PCR even when the samples were stored at temperatures greater than 4°C for up to 30 days2, 3, 4 & 5. Nasal swabs have also been collected in DNA/RNA Shield in Nigeria to successfully identify respiratory viral infections, including coronavirus6. Additionally, DNA/RNA Shield has been utilized to gather nasal swabs for surveying Middle East Respiratory Syndrome Coronavirus (MERS-CoV) infections among camels in the middle east using RT-qPCR7, due to its ability to inactivate pathogens for safe sample transportation. In addition to nasal swabs, a wide range of other sample types have also been collected in DNA/RNA Shield and the extracted DNA and RNA was more than fit for viral detection via PCR or Next-Gen Sequencing techniques; this includes conjunctival swabs7, epithelial tissue8, brain tissue9, prawn larvae10, feces11 & 12, Urine13, Ticks14, cell lines15, lower fornix swabs16, wasps and flies17, hair18, hemolymph19, mosquitoes20, oropharyngeal swabs21, liver tissue22, peripheral blood mononuclear cells22, and plasma23 & 24.
DNA/RNA Shield is also more than capable of inactivating a variety of microbes and viruses, making it well suited for handling and transporting biohazardous samples safely. A number of common human pathogens have been shown by Zymo Research to become completely inactivated when exposed to DNA/RNA Shield for 5 minutes (Figure 3).
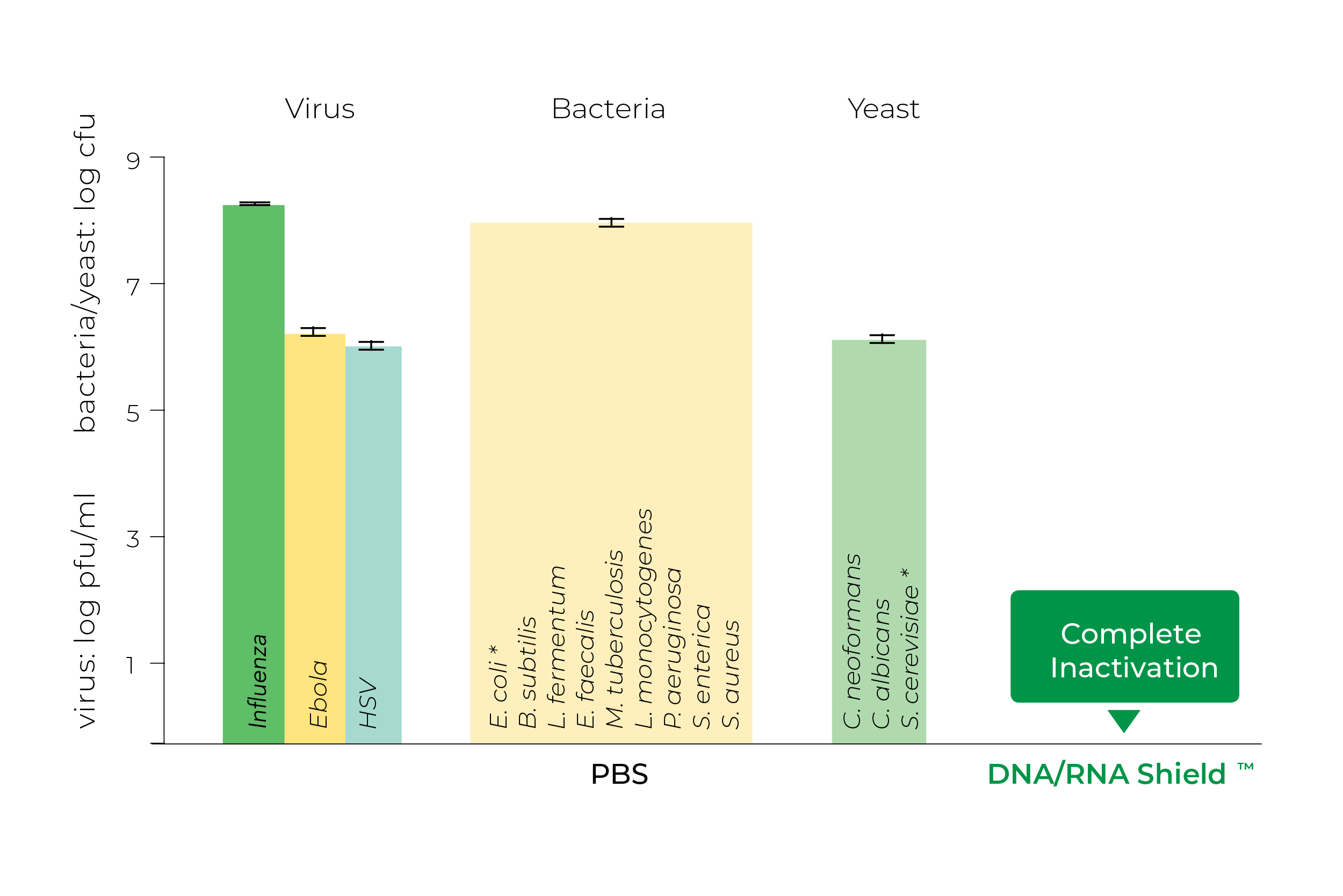
In addition, a study by Jaquelyn Horsington and colleagues in 2020 demonstrated that DNA/RNA Shield is far superior to RNAlater™ for inactivating foot and mouth disease virus8. In fact, several studies have used DNA/RNA Shield specifically for inactivating West Nile Virus5, 23, 25, & 32 and MERS-CoV7.
The compatibility of DNA/RNA Shield with Zymo Research’s popular nucleic acid extraction kits has already been independently validated in a number of studies, including the Quick-RNA Miniprep and Microprep Kits7, 9, 15, 19, 22, 24, 26, 27, 28, 29 & 30, ZymoBIOMICS DNA/RNA Miniprep Kit11 & 12 & 31, Quick-DNA/RNA Viral Kit32, Direct-zol RNA Miniprep Kits33 & 34, Quick-RNA Whole Blood kit1, Quick DNA/RNA Pathogen Miniprep Kit20, Quick-RNA Viral kit6, and Quick-DNA/RNA Miniprep Plus Kit14. However, DNA/RNA Shield has also already been used in conjunction with many other suppliers’ nucleic acid purification products to effectively extract high quality DNA and RNA, including the High Pure Viral RNA Kit from Roche10, the innuPREP RNA Mini Kit from Analytik Jena35, and Qiagen’s QIAamp® DNA Mini Kit36, QIAamp® Viral RNA Miniprep kit23, and QIAamp® MinElute Virus Spin Kit17. Tried-and-true standard TRIzol® RNA extraction has been shown to be compatible with DNA/RNA Shield as well9, 28 & 37.
Due to the high volume of specimens that need to be tested, it is also essential that both high-throughput and automated nucleic acid extraction platforms are compatible with the sample collection media. Direct compatibility alleviates the initial processing by reducing the number of steps and time. Samples in DNA/RNA Shield can be loaded into the sampling wells of the plate loaded into the automated platform without additional incubations or handling.
Zymo Research’s ZR-96 Genomic MagPrep and Qiagen’s PowerMag® Microbial DNA Extraction kit have been used successfully with samples in DNA/RNA Shield on the Tecan FreedomEVO platform®38. The NucleoMag® VET Kit from Macherey-Nagel has also extracted high quality viral RNA from samples in DNA/RNA Shield on ThermoFisher’s KingFisher™ Flex Magnetic Particle Processor8. Additionally, Qiagen’s EZ1® Virus Mini Kit v2.0 and EZ1® Advanced XL Instrument was used to extract RNA from samples stored in DNA/RNA Shield for RT-qPCR based viral detection13. Similarly, the MagNa Pure LC instrument and Magna Pure LC Total Nucleic Acid Isolation Kit from Roche also successfully extracted molecular biology grade viral RNA from samples in DNA/RNA Shield18. Biomerieux’s NucliSENS® Extraction kit and NucliSENS easyMAG® system has been used extensively with nasopharyngeal swabs stored in DNA/RNA Shield for total nucleic acid extraction and viral detection2, 3, 4, & 5. High quality RNA has also been extracted from samples in DNA/RNA Shield using Invitrogen’s iPrep™ PureLink™ Virus kit and iPrep™ Purification Instrument21 and Qiagen’s QIAamp® 96 DNA Blood Kit39. Remarkably, the QIAamp® Viral RNA Mini kit, MagNa PURE LC extraction system, the EZ1® Advanced XL system and Virus Kit, NucliSENS® easyMAG® extraction system, and KingFisher™ Flex Magnetic Particle Processor are already used in several COVID-19 molecular diagnostic workflows that have received emergency use authorization (EUA) by the FDA40.
DNA/RNA Shield’s ability to preserve RNA at ambient temperatures for extended periods of time and inactivate human pathogens, along with its compatibility with a variety of commercially available nucleic acid extraction products, make it an ideal sample collection solution for global COVID-19 surveillance. In fact, DNA/RNA Shield has already been widely adopted for sample collection by a number of government agencies and companies because of its superior storage capabilities, including the National Aeronautics and Space Administration (NASA), the Food and Drug Administration, United Stated Department of Agriculture, United States Department of Defense, LabCorp and uBiome.
Learn more about how DNA/RNA Shield can improve your COVID-19 testing workflow
References
- Zhang, J, et al., "Comparison of Three Different Pretreatment Methods for Blood RNA Extraction" Forensic Science International: Genetics Supplement Series (2019).
- Galanti, M, et al., "Rates of asymptomatic respiratory virus infection across age groups" Epidemiology & Infection, 147 (2019).
- Galanti, M, et al., "Longitudinal active sampling for respiratory viral infections across age groups" Influenza and Other Respiratory Viruses, 13(3): 226-232 (2019).
- Birger, R, et al., "Asymptomatic shedding of respiratory virus among an ambulatory population across seasons" mSphere, 3(4) (2018).
- Shaman, J, et al. "Asymptomatic summertime shedding of respiratory viruses" The Journal of Infectious Diseases 217(7): 1074-1077 (2018).
- Kolawole, O, et al., "Etiology of respiratory tract infections in the community and clinic in Ilorin, Nigeria" BMC research notes, 10(1): 712 (2017).
- Nowotny, N, Kolodziejek, J, “Middle East respiratory syndrome coronavirus (MERS-CoV) in dromedary camels, Oman, 2013” Euro Surveillance, 19(16) (2014).
- Horsington , J, et al., "Inactivation of foot and mouth disease virus in epithelium samples for safe transport and processing in low containment laboratories" Journal of Virological methods, 276:113770 (2020).
- Brunker, K, et al., "Rabies elimination programmes [version 1]” peer review: awaiting.
- Pasookhush, P, et al., "Transcriptomic analysis of Macrobrachium rosenbergii (giant fresh water prawn) post-larvae in response to M. rosenbergiinodavirus (MrNV) infection: de novo assembly and functional annotation" BMC Genomics, 20(1): 762 (2019).
- Wang, Z, "Comparing Gut Microbiome and Viromein the Breast Milk-and Formula-fed Late Preterm Infants" Electronic Theses and Dissertations, 3375 (2019). https://openprairie.sdstate.edu/etd/3375
- Chard, A, et al., "Associations between soil-transmitted helminthiasis and viral, bacterial, and protozoal enteroinfections: a cross-sectional study in rural Laos" Parasites & Vectors, 12(1): 216 (2019).
- Tan, Susanna K., et al. "Impact of Pretransplant Donor BK Viruriai Kidney Transplant Recipients" The Journal of infectious diseases, 3 (2019).
- Mendell, N, et al., "Detection of Rickettsiae, Borreliae, and Ehrlichiaein Ticks Collected from Walker County, Texas, 2017–2018" Insects, 10(10): 315 (2019).
- Jiwaji, Meesbah, et al. "Providence virus: An animal virus that replicates in plants or a plant virus that infects and replicates in animal cells?" PLoS ONE, 14(6) (2019).
- Lalitha, P, et al., "Unbiased Pathogen Detection and Host Gene Profiling for Conjunctivitis" Ophthalmology (2019).
- Bennett, A, et al., "Diverse RNA viruses of arthropod origin in the blood of fruit bats suggest a link between bat and arthropod viromes" Virology, 528: 64-72 (2019).
- Spiri, A, et al., "Environmental Contamination and Hygienic Measures After Feline Calicivirus Field Strain Infections of Cats in a Research Facility" Viruses, 11(10): 958 (2019.),
- Grandjean, F, et al., "A new bunya-like virus associated with mass mortality of white-clawed crayfish in the wild" Virology, 533: 115-124 (2019).
- Hepp, C, et al., "Phylogenetic analysis of West Nile Virus in Maricopa County, Arizona: Evidence for dynamic behavior of strains in two major lineages in the American Southwest" PLoS ONE, 13(11) (2018).
- Šimić, I, et al., "Molecular and serological survey of lyssaviruses in Croatian bat populations" BMC Veterinary Research, 14(1): 274 (2018).
- Elmasry, S, et al. "Detection of occult hepatitis C virus infection in patients who achieved a sustained virologic response to direct-acting antiviral agents for recurrent infection after liver transplantation" Gastroenterology, 152(3): 550-553 (2017).
- KolodziejekJ, S, et al., “West Nile Virus Positive Blood Donation and Subsequent Antomological Investigation, Austria, 2014” PLoS ONE, 10(5) (2015).
- Phommanivong, V, et al., “Co-circulation of the dengue with chikungunya virus during the 2013 outbreak in the southern part of Lao PDR” Trop Med Health (2016).
- Knutie, S, and Gotanda, K, "A non-invasive method to collect fecal samples from wild birds for microbiome studies" Microbial Ecology, 76(4): 851-855 (2018).
- Ishida, Y, et al., "Hepatic IFN-Induced Protein with Tetratricopeptide Repeats Regulation of HCV Infection" Journal of Interferon & Cytokine Research, 39(3): 133-146 (2019).
- Gardner, J, and Herbst-Kralovetz, M, "IL-36γ induces a transient HSV-2 resistant environment that protects against genital disease and pathogenesis" Cytokine, 111: 6371 (2018).
- Elbadry, M, et al., “High prevalence of asymptomatic malaria infections: a cross sectional study in rural areas in six departments in Haiti” Malaria Journal, 14:510 (2015).
- Gardner, J, et al., "IL 36γ Is a Key Regulator of Neutrophil Infiltration in the Vaginal Microenvironment and Limits Neuroinvasion in Genital HSV 2 Infection" The Journal of Immunology (2019).
- Dornfeld , D, et al. "SMARCA2 regulated host cell factors are required for MxA restriction of influenza A viruses." Scientific Reports, 8(1): 2092 (2018).
- Stamps, B, et al., "Characterization of the microbiome at the world’s largest potable water reuse facility" Frontiers in Microbiology, 9: 2435 (2018).
- Ronca , S, et al., "Phenotypic and Genotypic Characterization of West Nile Virus Isolate 2004Hou3" International Journal of Molecular Sciences, 20(8):1936 (2019).
- Retallack , H, et al., "Genome Sequence of a Divergent Avian Metapneumovirus from a Monk Parakeet Myiopsitta monachus” Microbiology Resource Announcements, 8(16) (2019.
- Bryant, W, et al., "Small RNA-Seq Analysis Reveals miRNA Expression Dynamics Across Tissues in the Malaria Vector, Anopheles gambiae" G3: Genes, Genomes, Genetics, 9(5): 1507-1517 (2019).
- Littwitz-Salomon, E, et al. "The cytotoxic activity of natural killer cells is suppressed by IL-10+ regulatory T cells during acute retroviral infection" Frontiers in Immunology, 9: 1947 (2018).
- Zainabadi, K, et al., "A novel method for extracting nucleic acids from dried blood spots for ultrasensitive detection of low-density Plasmodium falciparum and Plasmodium vivax infections" Malaria Journal, 16(1):377 (2017).
- Carrau , L, et al. "Chikungunya virus vaccine candidates with decreased mutational robustness are attenuated in vivo and have compromised transmissibility" Journal of Virology, 93(18) (2019).
- Hugerth, L, et al. “A comprehensive automated pipeline for human microbiome sampling, 16S rRNA gene sequencing and bioinformatics processing” bioRxiv, doi: https://doi.org/10.1101/286526 (2018).
- Adams, M, et al., “An ultrasensitive reverse transcription polymerase chain reaction assay to detect asymptomatic low-density Plasmodium falciparum and Plasmodium vivax infections in small volume blood samples” Malaria Journal, 14:520 (2015.
- https://www.fda.gov/medical-devices/emergency-situations-medical-devices/emergency-use-authorizations#coronavirus2019
- https://coronavirus.jhu.edu/map.html


