Sample Preservation: The Origin of Bias
As one of the most powerful and prevalent tools for developing gene expression profiles, RNA-sequencing (RNA-Seq) is vulnerable to biases. The instability of DNA and RNA at ambient temperatures means researchers must be especially careful when collecting samples in order to ensure no loss of sample information prior to nucleic acid extraction. One common method for stabilizing nucleic acid in biological samples is to flash-freeze the samples in liquid nitrogen. While effective, flash freezing can be overly cumbersome and impractical for outside of a controlled lab environment. As a result, some researchers turn to commercially available reagents such as RNAlaterTM to stabilize nucleic acid at ambient temperature.
A recent study from researchers at the University of Minnesota shows that storing a biological sample in RNAlaterTM could have a significant impact on the gene expression profiles when compared to samples flash frozen in liquid nitrogen. The researchers found 2,708 genes were shown to be differentially expressed when compared to samples stored in RNAlater™ and those that were flash frozen, with some of this bias attributed to the GC content of the genes, stating
“…we recommend that researchers use caution when comparing gene expression values derived from RNAseq datasets that may have variable storage conditions” (Passow et al.)
The significance of liquid biopsies is their ability to detect early disease with just a simple blood draw. As technologies improve the detection of specific disease biomarkers, earlier adoption of more effective targeted treatments is possible. This potential has been gaining a lot of attention, leading to significant investments by companies like Illumina and Roche.
For researchers who rely on RNAlater™ for preserving their RNA, this has huge implications that are concerning to those using RNA-Seq to develop accurate gene expression profiles. Biased changes to the observed expression profile can greatly influence the final data and cause changes in the interpretation of results.
Ideally, samples should be protected from the effects of freeze/thaw and maintain expression profiles from the time of collection (Figure 1). This requires that the collection and preservation method(s) protect against bias under all conditions.
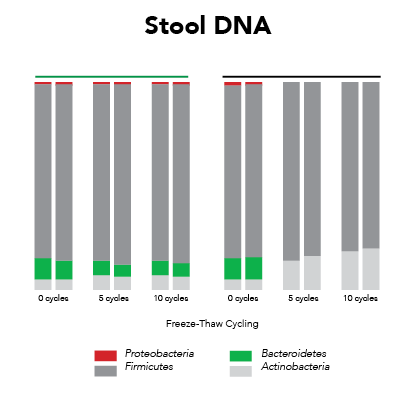
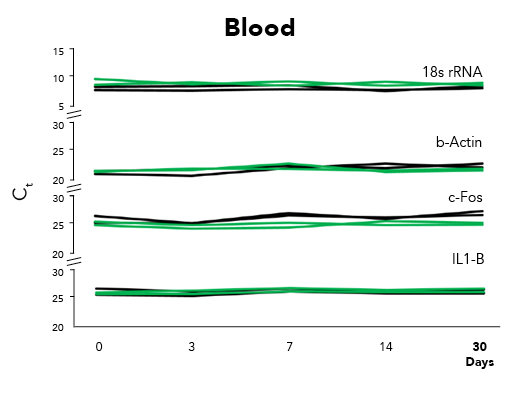
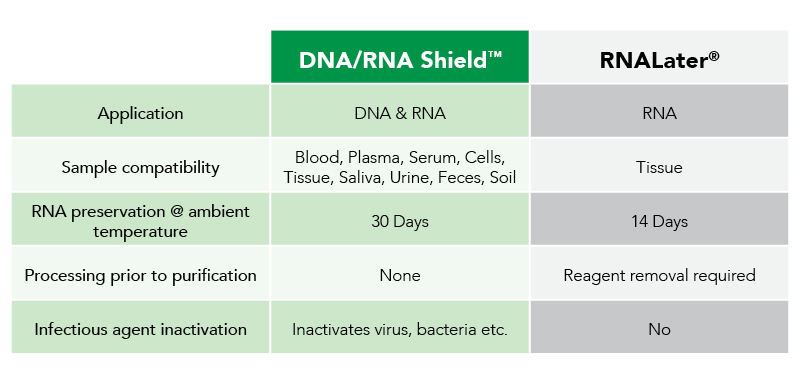
Fortunately, Zymo Research has eliminated these biases by the development of DNA/RNA Shield™. DNA/RNA Shield is a preservative that maintains the samples expression profiles of biological samples at both ambient and frozen temperatures (Figure 2). Additionally, DNA/RNA Shield protects both DNA and RNA from multiple samples types, which is not observed for RNALater™ (Figure 3).
As research studies grow in complexity and analytical techniques increase in sensitivity, the influence of sample collection and storage on data will be more often considered. Moreover, use of non-optimized technologies can lead to biased expression profiles, as recognized by Passow and colleagues in RNAlater and flash freezing. Therefore, next-generation technologies like DNA/RNA Shield are on track to become the new standard for sample collection, transportation and storage by enabling unbiased data collection.
References:
Passow, Courtney N., et al. "RNAlater and flash freezing storage methods nonrandomly influence observed gene expression in RNAseq experiments." bioRxiv (2018): 379834.


