What is TRIzol®?
A BREAKDOWN OF TRIZOL®AND HOW TO OPTIMIZE YOUR RNA EXTRACTION
TRIzol® - the Gold (Pink) Standard
RNA extraction from TRIzol® has long been considered the “gold standard” for RNA purification. So much so that other commercial companies offer similar reagents with their own nomenclature (TRI Reagent®, RNAzol®, QIAzol®, TriPure, TriSure, etc.). The all-in-one reagent offers excellent capability to lyse a variety of complex samples. TRIzol® is easily scalable for accommodating very large inputs, recovers ample RNA yield and is also an effective inactivator of RNases, preventing degradation of samples during purification 1. Overall, TRIzol® preserves the RNA quality, integrity and quantity, allowing it to double as a storage medium as well as an extraction/purification solution.
However, there are major obstacles in the laborious method such as the use of chloroform, long centrifugation times and the need for precipitation steps during the isolation process 2. The procedure can be cumbersome and require technical skill to ensure adequate recovery of RNA from samples.
How does TRIzol® work?
TRIzol® reagent is an acid-guanidinium-phenol based reagent ideally designed for the extraction of RNA (as well as DNA and protein) from various biological sample inputs. The low pH (acidic) of TRIzol® controls to separate RNA from DNA and protein, while a high pH can cause RNA and DNA to be isolated together. The guanidinium salt serves as a chaotropic agent to denature proteins and the phenol (commonly indicated as a pink color) is an organic compound also used to extract nucleic acids and proteins 3,4.
After solubilization and homogenization of samples in TRIzol®, the RNA, DNA and protein are differentially extracted by the addition of a phase separation reagent (chloroform, BCP or BAN). The solution separates the RNA away from DNA and protein into different layers (Figure 1). An upper, clear aqueous phase mainly contains RNA, and the middle interphase and lower organic phase contains DNA, proteins and lipids 3. Subsequently, the RNA in the upper aqueous phase is then collected by alcohol-based precipitation. After lengthy incubation and centrifugation steps, the resulting white pellet (if any) is then washed in an ethanol solution, air-dried and resuspended in the final sample buffer.
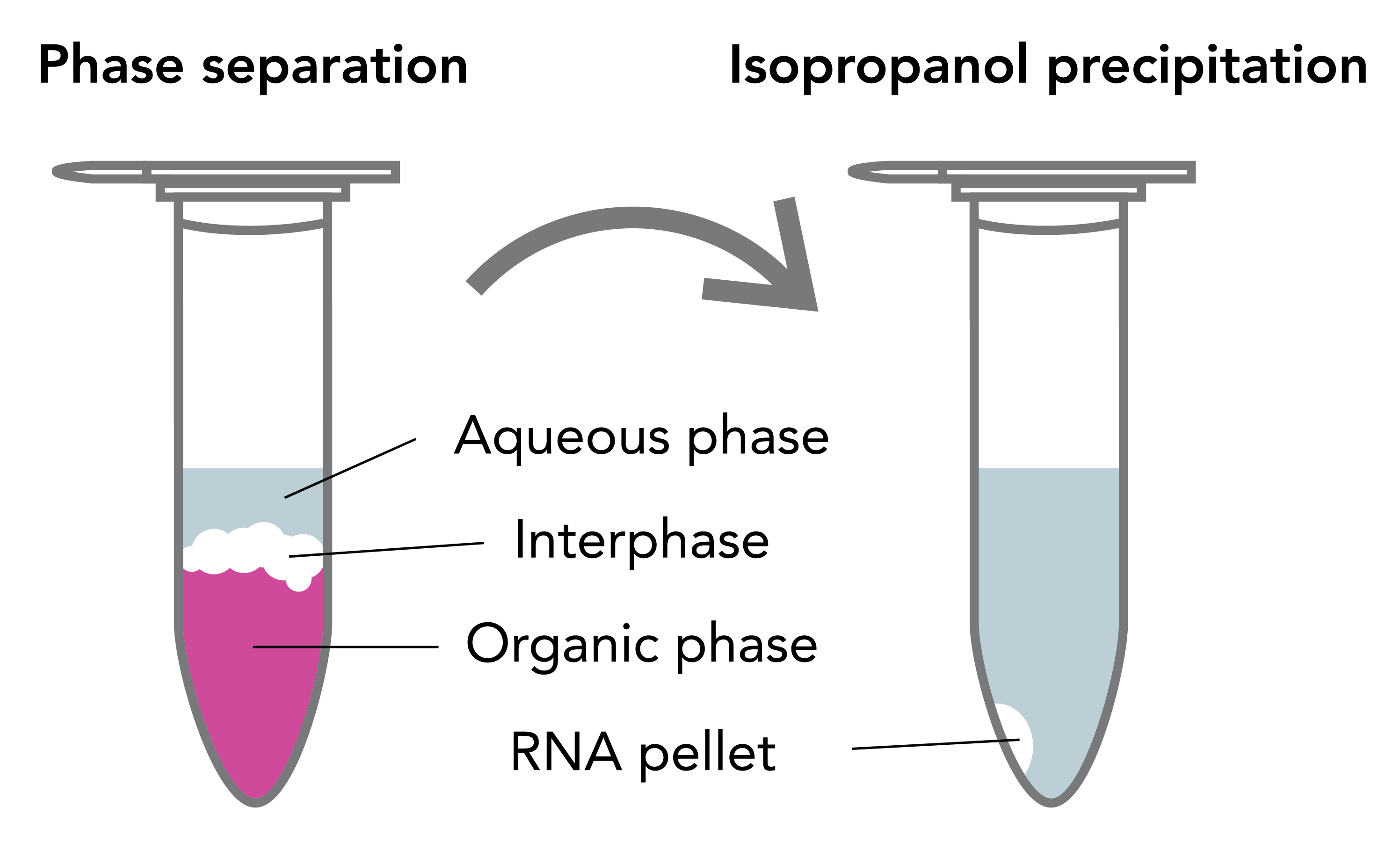
TRIzol® RNA Extraction and Common Issues
Although TRIzol® is known to be a powerful reagent in itself, adding TRIzol® to a sample is not a complete cure-all. One must consider the type of sample (biological liquids, cells or solid tissue) and the volume of TRIzol® used. If the upstream processing is not well thought out, samples can result in incomplete lysis and DNA- or phenol-contamination. Also, a bias in micro/small RNA (17-200 nt) and large RNA (>200 nt) can occur. Lastly, the traditional RNA extraction can be tedious when performing multiple samples, which makes the method unadaptable to high-throughput automated liquid handling platforms.
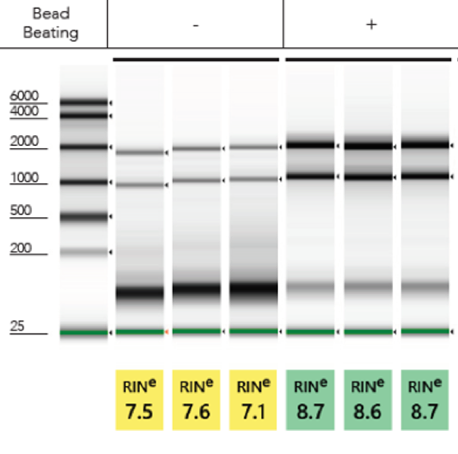
Inefficient Lysis:To efficiently lyse a sample, the TRIzol® to sample volume ratio is very important. More often than not, increasing the volume of TRIzol® than what the traditional method recommends tends to solve most issues with RNA extraction. Alternatively, performing mechanical lysis in TRIzol® prior to purification via bead beating tubes and a high-speed homogenizer will yield robust total RNA (Figure 2). Incorporating both mechanical homogenization and increasing the volume of TRIzol® will ensure maximum quality RNA.
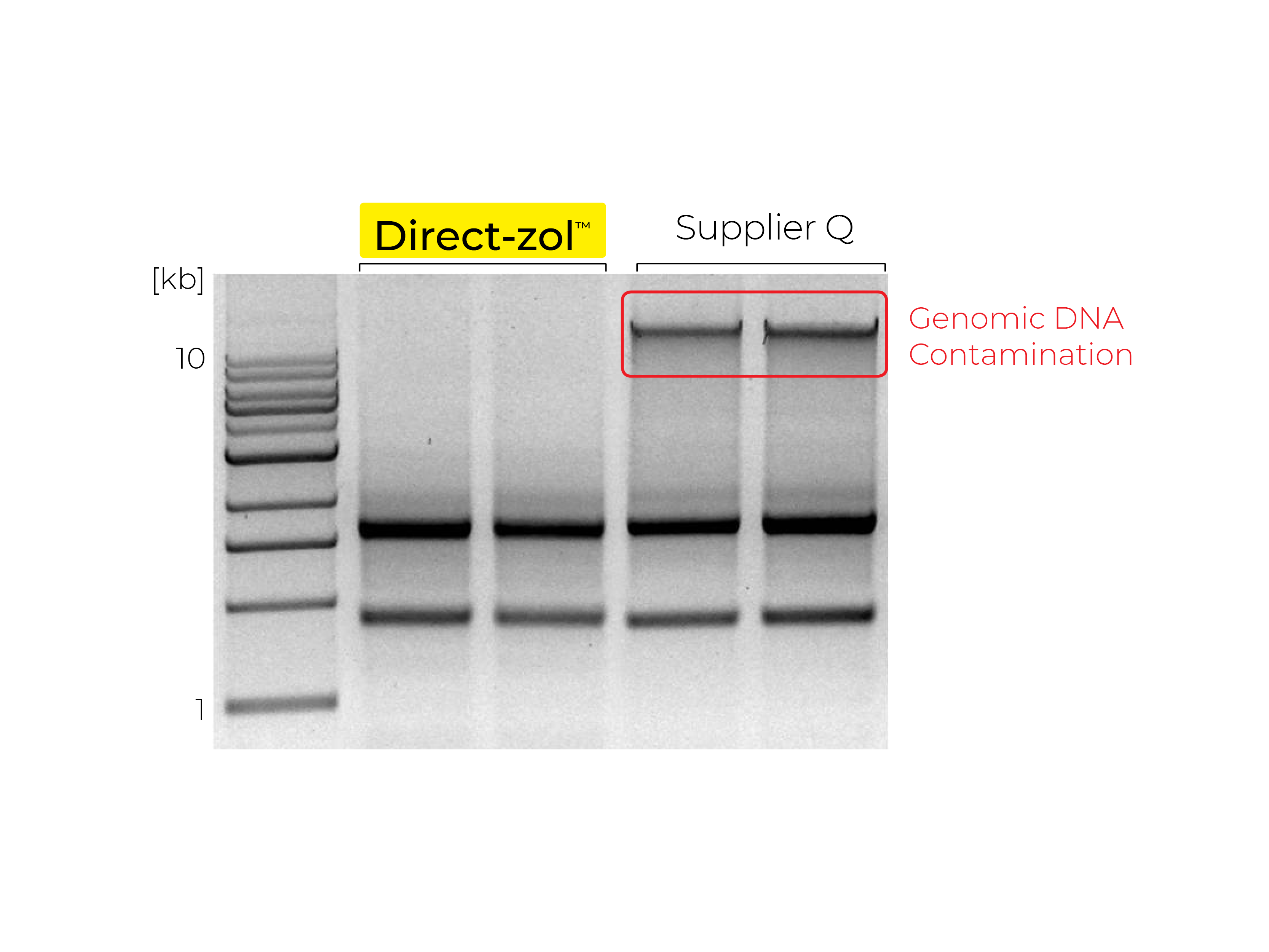
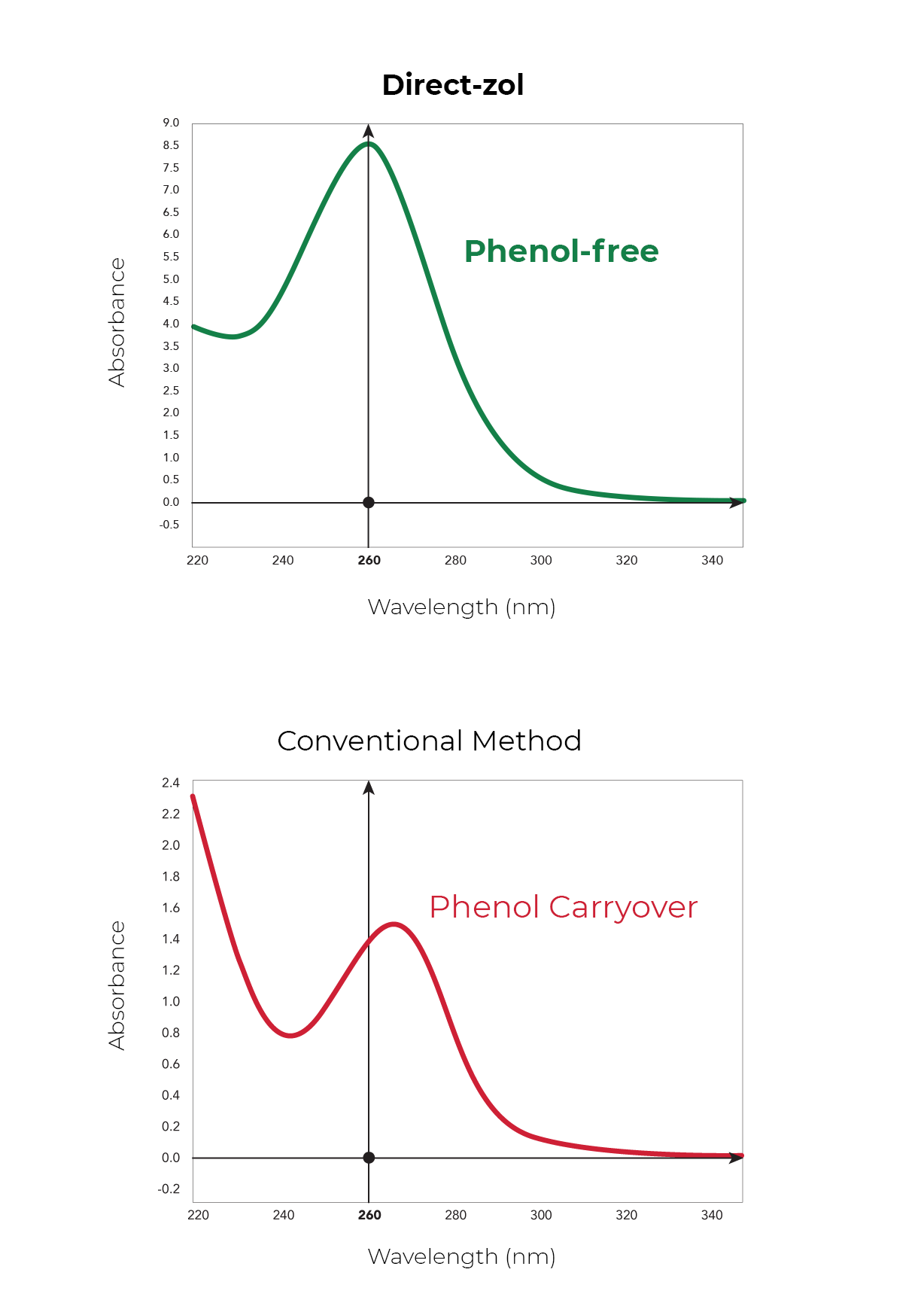
Phenol and Genomic DNA Contamination: During the removal of the upper aqueous phase from the inter- and organic phase, cross-contamination can occur. Organic phenol, protein, lipids and DNA can be unintentionally lifted into the aqueous phase causing low quality RNA (Figure 3). To determine phenol contamination, a simple test can be seen when quantifying an RNA sample on a spectrophotometer, used for the measurement of transmittance or reflectance of solutions. An abnormal, absorbance maxima at 270 nm indicates phenol contamination (Figure 4). With genomic DNA contamination, indicators can be inflated yields on the spectrophotometer and high molecular weight band(s) or a smear on an agarose gel using electrophoresis (Figure 5).
Beyond TRIzol®
With many non-organic and commercial kits that now exist for RNA purification, there are still labs today that wish to continue to use TRIzol®. Some may have samples stored in TRIzol® long-term or just prefer the reagent for purification. However, there is a greater need for faster handling time, reliability and consistency for ultra-pure RNA.
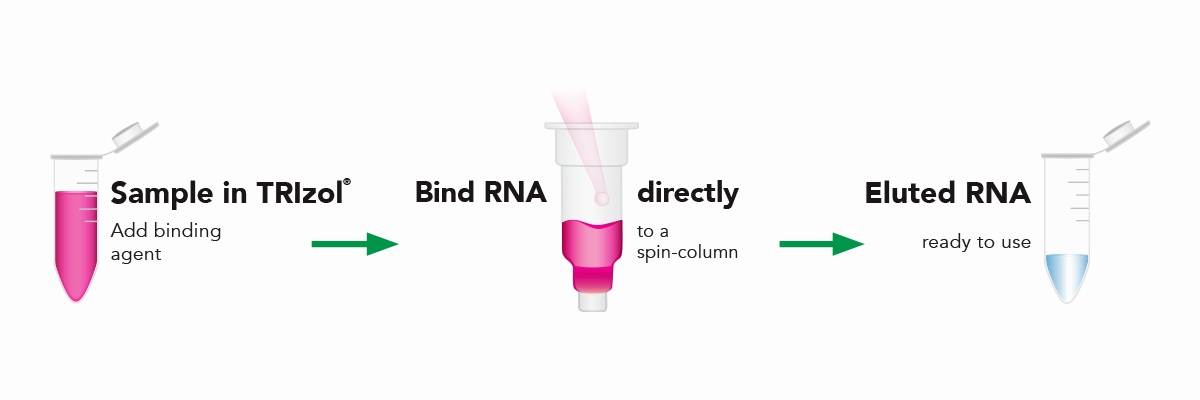
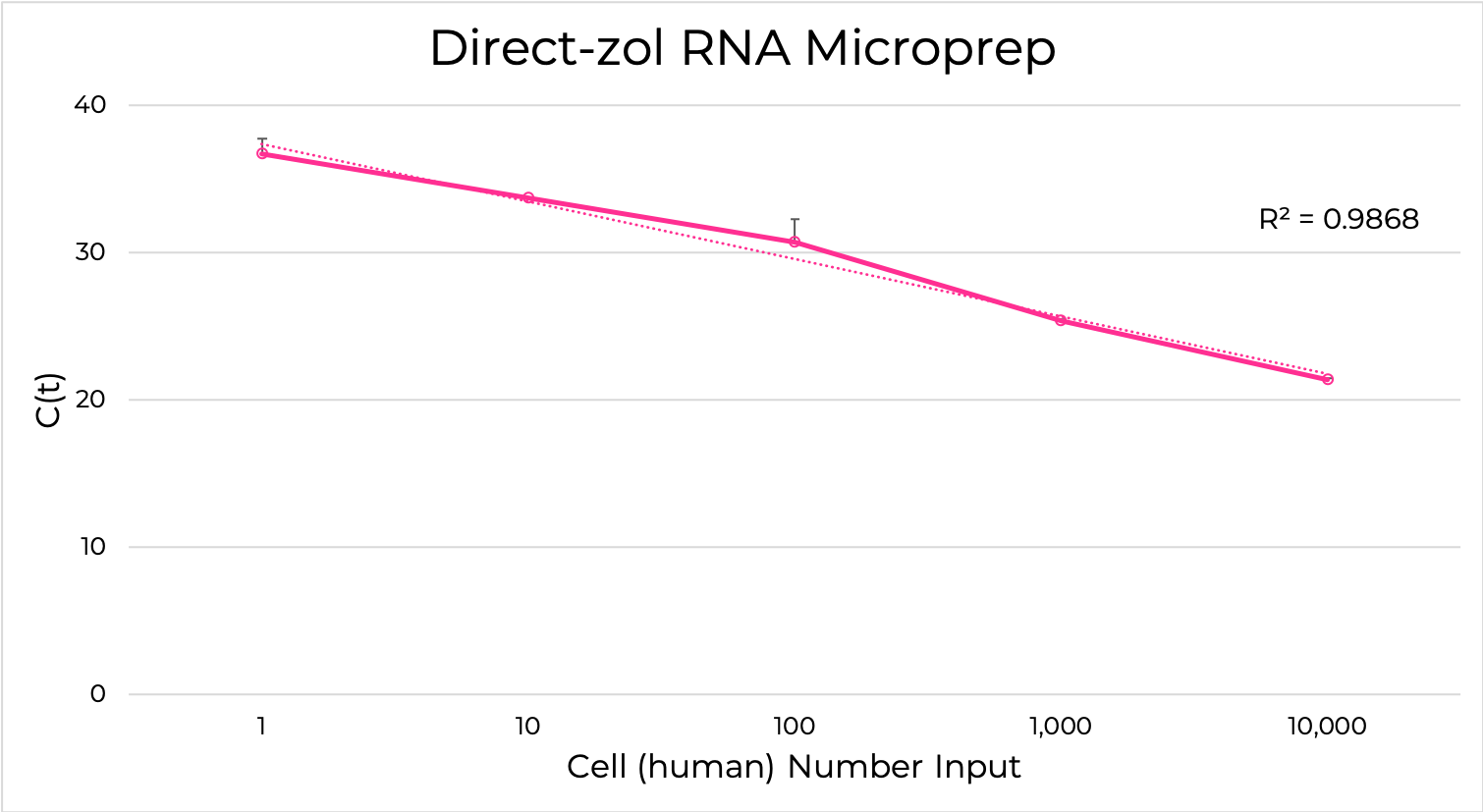
The Direct-zol RNA kits are designed to purify any sample in TRIzol® without DNA or phenol contamination and tedious phase-separation or precipitation steps. Simply add the TRIzol® lysate mixed with ethanol to the binding column, wash and elute (Figure 6). The resulting RNA is ready for sensitive downstream applications such as RT-qPCR and Next-Gen Sequencing. Furthermore, the Zymo-Spin silica-based binding column is uniquely designed to capture unbiased total RNA down to picogram amounts (single-cell) (Figure 7).
Try a free sample of the Direct-zol RNA kit
Citations:
[1] Beintema JJ, van der Laan JM "Comparison of the structure of turtle pancreatic ribonuclease with those of mammalian ribonucleases". 1986.
[2] Material safety data sheet from Molecular Research Center, Inc. (MRC).
[3] Brawerman G, Mendecki J, Lee Sy. A procedure for the isolation of mammalian messenger ribonucleic acid. Biochemistry. 1972.
[4] Chomczynski P et al. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987.