Score More Spore DNA
When thriving bacterial cells are threatened by harsh conditions and nutritional restrictions, the extreme stress can drive them into survival mode, forming endospores. These bacterial spores can be exceptionally tough, remaining viable even after exposure to extreme conditions due to their resistance to enzymatic lysis, desiccation, radiation, high temperatures, and chemical treatments such as disinfectants and denaturants. These resistances can be very challenging to metagenomic studies, as efficient lysis of spores is important to ensure an accurate representation of the microbial profile of a sample. Inefficient lysis of spores can lead to misrepresentation of the microbial profile and very little to no DNA recovery from the spores.
Why so difficult?
The extreme hardiness of spores and their resistance to lysis is a product of many factors. The outer most coating of the endospore is a proteinous layer that provides substantial enzymatic and chemical resistance. Beneath this layer are structures that provide further physical and chemical barriers: a peptidoglycan cell wall called the cortex, followed by a germ cell wall, and an inner membrane. Within this inner membrane is the core, which contains the DNA and ribosomes, and includes additional protective elements including dipicolinic acid and proteins that further protect the DNA from radiation and chemical damage.
Breaking it down
Spores are notably difficult to lyse, and heat treatments tend to be ineffective at liberating DNA from endospores. Additionally, enzymatic methods are dependent on an organism’s lytic susceptibility and the complex structure of spores tends to make them highly resistant even if the vegetative bacterium was susceptible. Mechanical lysis is widely accepted as the most effective method of lysing endospores as it is more robust and less specific.
To determine the efficiency of different lysis mechanisms on spores, Zymo Research compared three methods of lysis on Bacillus subtilis endospores: chemical, mechanical, and heat. The results indicate that the only way to achieve complete, unbiased lysis of bacterial endospores is to use high-density bead-beating beads with high speed or low speed homogenizers.
Spore Induction and Indirect Quantification
Bacterial spore formation was induced by inoculating Bacillus subtilis cells growth/sporulating medium with Bacillus subtilis and incubating for several days. Successful bacterial spore formation was determined by using Schaeffer and Fulton Spore Stain Kit (Sigma Aldrich) and viewed by microscopy. Bacterial spores were then concentrated and frozen in a PBS/ glycerol solution. Cell counting was not possible due to the extremely small size of the bacterial spores, so indirect counting was performed via plating serial dilutions of bacterial spore suspension.
Methods Surveyed
Evaluating the Efficacy of Chemical Lysis on Spores
Chemical lysis was attempted using several common commercially available lysis buffers (e.g. Zymo Research Genomic Lysis Buffer, Qiagen Buffer AL, and Qiagen Buffer AVL) which led to no significant DNA recovery, as anticipated (data not shown). This does not reflect a comprehensive review of chemicals with the potential to lyse endospores, it is just an evaluation of some of the most used cellular lysis buffers.
Evaluating the Efficacy of Thermal Lysis on Spores
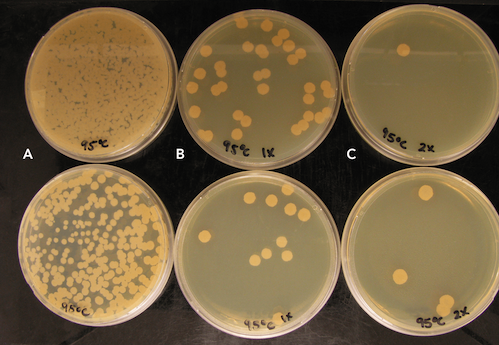
Bacillus subtilis spores were treated at room temperature (RT), 55 ˚C, 75 ˚C, and 95 ˚C for 1 hour. In all instances no substantial quantities of DNA were recovered in the purification, however spores treated at 95 ˚C for 1 hour experienced 106 decrease in cell growth when plated, in comparison to other spore treatments (Figure 1).
Evaluating the Efficacy of Mechanical Lysis on Spores
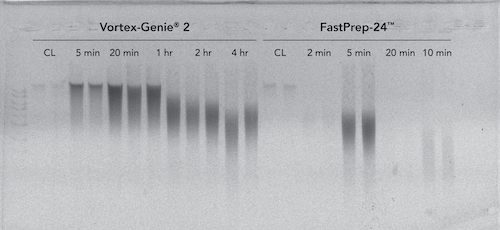
The ZymoBIOMICS DNA Miniprep Kit which contains a mixture of high density BashingBeads™ (0.1 and 0.5 mm) was evaluated in the context of two different types of homogenization systems, classified as high speed and low speed. It was found that the ZymoBIOMICS DNA Miniprep kit effectively lysed Bacillus subtilis endospores using a low speed homogenizer after 20 minutes (3,200 rpm; Vortex-Genie® 2) and using a high-speed homogenizer after 5 minutes (1 minute interval at 6.5 m/s with 5 minutes rest; FastPrep-24™) (Figure 2).
Increased bead-beating duration beyond 20 minutes on the low-speed device resulted in negligible changes in yield and minimal DNA loss/shearing. However, increased bead-beating duration beyond 5 minutes on the high-speed device resulted in substantial DNA degradation and loss of DNA. It is of note, that the high-speed device generated significant heat within the ZR BashingBead™ Lysis Tubes, which may have been a cause of the substantial DNA degradation.
Mechanical Lysis Comparison
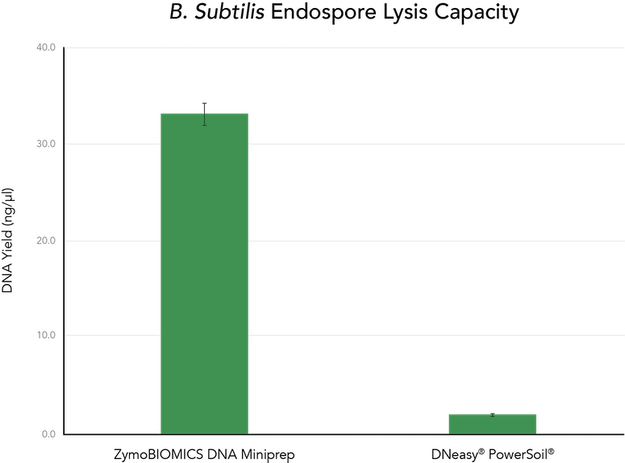
The ZymoBIOMICS DNA Miniprep was compared to the DNeasy® PowerSoil® (Qiagen) kit to evaluate the lysis efficiency and recovery of DNA from solutions containing 6.0 x 108 CFU of B. subtilis endospores. The ZymoBIOMICS DNA Miniprep was consistently able to lyse the B. subtilis endospores and recover the DNA, while the DNeasy® PowerSoil® kit was incapable of recovering measurable quantities of DNA (Figure 3).
Protocol for DNA Isolation from Spores
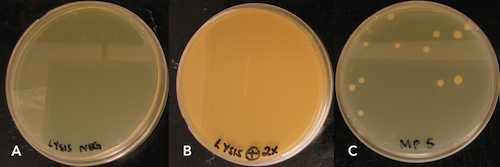
The ZymoBIOMICS DNA Miniprep’s high density bead formulation (0.1 & 0.5 mm) was shown to be effective in lysing B. subtilis endospores with high and low speed disruptors indicating the versatility of the kit. Furthermore, lysis efficiency was determined to be greater than 99% as determined by plating spore lysates (Figure 4). The number of viable colony forming units plated after lysis was minimal and equated to picograms of unrecoverable DNA, suggesting that lysis using high density BashingBeads™ (0.1 and 0.5 mm) was nearly 100% efficient.
Protocol for how to extract DNA from spores:
- Add endospore sample/sample containing endospores to a ZR BashingBead Lysis Tube.
- Add 750 uL DNA/RNA Shield or ZymoBIOMICS Lysis Solution to the sample tube and cap tightly.
- Bead beat sample with a validated method for complete lysis of microbes, including spores:
- Low Speed: Vortex-Genie®
- Place tube on horizontal tube adapter
- Turn Vortex-Genie® on at maximum speed
- Bead beat for 40 minutes (20 minutes is sufficient for spore lysis, however 40 minutes will ensure complete lysis of microbes for metagenomic studies)
- High Speed: Fastprep-24™
- Place tube in bead beater and secure
- Bead beat at maximum speed (6.5 m/s) for 1 minute
- Allow sample to rest for 5 minutes. Insufficient rest may result in system error.
- Repeat 5 times for a total of 5 minutes of bead beating
- Low Speed: Vortex-Genie®
- Spin sample at >10,000 g for 1 minute
- Process the supernatant using the ZymoBIOMICS DNA Kit


