How to Extract RNA from Hard-to-Lyse Samples in Trizol®
Learn the best extraction methods for challenging sample types
The Challenge of RNA Isolation
RNA isolation is a challenging procedure. Unlike DNA, which is highly stable, RNA is very unstable, quickly digested by RNase enzymes if their activity isn’t immediately inhibited. Traditional RNA isolation protocols are also tedious, time-consuming, and unscalable. Since RNA isolation is critical to many scientific enterprises, researchers have optimized protocols and reagents to speed up and simplify the process.
One popular reagent in RNA isolation protocols is TRIzol®. Composed of phenol and guanidine thiocyanate in a mono-phase solution, TRIzol® (or TRI Reagent®) is a cell lysis and RNase-inhibiting solution used for the simultaneous isolation of RNA, DNA, and proteins from biological samples. The components of TRI Reagent® facilitate immediate and effective inhibition of RNase activity and isolate RNA from human, animal, plant, yeast, bacterial, and viral samples with consistent performance on all quantities of tissues or cultured cells.
TRIzol® for Isolating RNA from a Range of Samples
TRIzol® was specifically formulated to isolate total RNA from any sample type in the modern research laboratory, whether in academia or industry: cells, hard to lyse or solid tissue, and infectious biological samples.
Cells
TRIzol® permits efficient RNA isolation from cells, because it quickly breaks down cell structures and inactivates RNases in the sample, protecting RNA from degradation.
Hard-to-Lyse Samples or Solid Tissue
The chemical makeup of TRIzol® also facilitates the release of RNA from hard-to-lyse cells and tissue samples with the aid of mechanical homogenization and/or Proteinase K treatment.
Infectious Biological Samples
TRIzol® inactivates pathogens in plasma, serum, stool, blood, and other biological sample types, eliminating infection risk for researchers and simplifying RNA extraction.
Tips for Efficient RNA Extraction with TRIzol®
RNA extraction with TRIzol® combines phenol and guanidine thiocyanate in a mono-phase solution, facilitating immediate and effective inhibition of RNase activity. However, when preparing biological samples for RNA isolation, certain considerations should be taken into account for an optimal outcome. Here are some sample preparation tips to ensure efficient RNA extraction from cells, hard-to-lyse/solid tissue, and infectious biological samples:
Cells
Due to their cellular structure, mammalian cells are the easiest to lyse in TRIzol®, typically needing no additional chemical lysis or mechanical homogenization. To maximize RNA isolation efficiency, adhere to the following while preparing cells in TRIzol®:
- Add 1 ml TRIzol® to 5-10 x 106 fresh, pelleted mammalian cells. Pipette up and down until cells are fully lysed and homogenous.
- Tip 1: If the lysate remains cloudy, opaque, and/or viscous, increase the volume of TRIzol® until clear.
- Tip 2: After lysis with TRIzol®, centrifuge the lysate at max speed for 1-2 minutes and transfer the supernatant into a new tube prior to processing. Leave at least 50 μl at the bottom of the old tube to avoid transferring part of the pellet.
Hard-to-lyse/solid tissue and infectious biological samples
Solid tissues, including heart, lung, liver, muscle, and cartilage, are more difficult to lyse than cells. Additionally, gram-positive bacteria and yeast cells (ex: Bacillus, Listeria, Saccharomyces) are also considered hard-to-lyse due to polysaccharides in their cell walls.
Infectious biological samples, such as plasma, serum, stool, and whole blood, are complex, highly proteinated samples with a mixture of easy and tough-to-lyse components, and therefore should be handled like hard-to-lyse samples. To maximize RNA isolation efficiency, adhere to the following while preparing tough-to-lyse samples, solid tissue, and infectious biological samples in TRIzol®:
-
Tip 1: Unlike cells, these sample types require additional chemical lysis and/or mechanical homogenization.
- We recommend mechanical homogenization as the optimal approach for lysing these samples. For high-speed homogenization (e.g., Bertin Precellys), add 1 ml TRIzol® to a fresh tissue sample (10% w/v) and bead beat at maximum speed for 30-60 seconds. For low-speed homogenization (e.g., Disruptor Genie), bead beat at maximum speed for 5-10 minutes. The exact amount of time is sample type-dependent.
- Chemical lysis with Proteinase K treatment can also be performed, either with or without mechanical homogenization. If mechanical homogenization was performed, treat the sample in Proteinase K for 30 minutes. If not, increase Proteinase K treatment time to 2-5 hours.
- Tip 2: After homogenization and/or Proteinase K treatment, centrifuge down any debris and transfer the supernatant into a new tube prior to purification.
- Tip 3: If the supernatant/lysate is still cloudy, opaque and/or viscous, increase the volume of TRIzol® until clear.
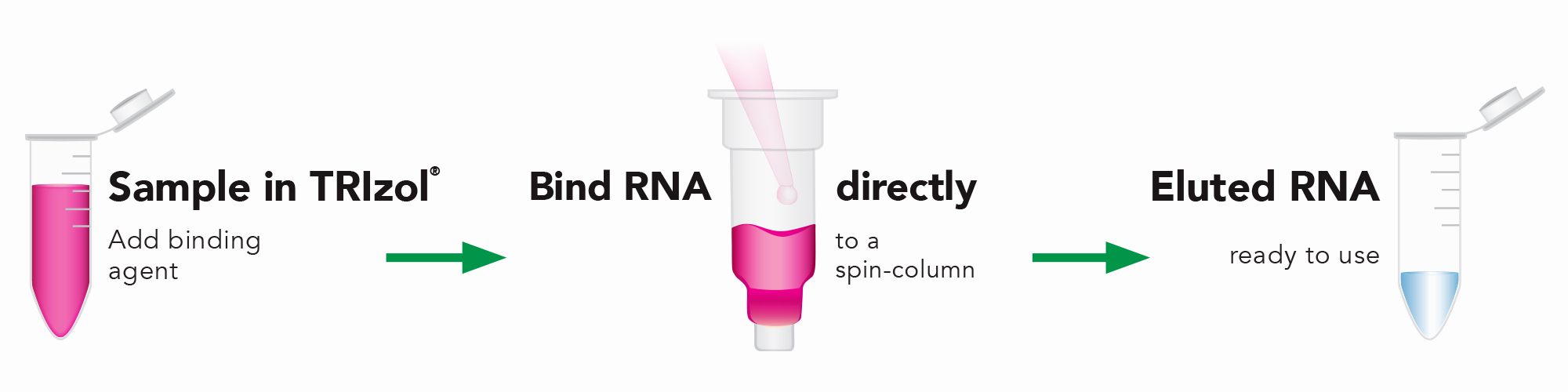
Extract RNA from TRIzol in 7 Minutes with Direct-zol RNA Kits
Traditional TRIzol® RNA extraction is an elaborate, time-consuming protocol. Once cells are lysed, then the homogenate must be phase-separated by adding bromochloropropane or chloroform and centrifuging the sample. After centrifugation, three phases are visible: the aqueous phase (RNA), the organic phase (DNA), and the interphase (proteins).
RNA is precipitated from the aqueous phase by adding isopropanol, washing with ethanol, and solubilizing the final RNA pellet. Each step in this process is tedious and time consuming, taking more than an hour for the entire process, and must be performed with care. RNA yields and purity are often compromised due to incomplete cell lysis, contamination of the aqueous phase by the phenol phase, or incompletely dissolved final RNA pellets.
The Direct-zol RNA kits by Zymo Research, designed to work with TRIzol®, TRI Reagent®, or similar acid-guanidinium-phenol based reagent, achieves total RNA extraction, including small/miRNAs, from any sample in only seven minutes. With the Direct-zol RNA kit, you’ll achieve consistent results, higher RNA yields (up to four-fold more), and a quicker processing time compared to the conventional RNA isolation method. No chloroform or phase separation is necessary, and there are no precipitation steps, eliminating aqueous phase contamination and incompletely dissolved RNA pellets.
To isolate RNA with the Direct-zol RNA kit, simply add sample lysed in TRIzol® directly to the Zymo-Spin Column and then bind, wash, and elute the RNA. The resulting DNA-free RNA is ready for any downstream application, including next-generation sequencing (NGS), RT/qPCR, northern blots, and other protocols requiring pure RNA.
The Direct-zol method can also be scaled up and automated with the Direct-zol-96 MagBead RNA kit, which is compatible with any robotic sample processor (e.g., Hamilton, Kingfisher, and Tecan) — making high-throughput, rapid extraction of high-quality RNA accessible to any research laboratory.
The Experience of Rapid, Efficient RNA Extraction
RNA extraction doesn’t have to be a dreaded part of your scientific protocols. The Direct-zol RNA kit makes RNA isolation from any sample type — including hard to lyse/solid tissue samples — a breeze. Due to its unsurpassed speed and ability to isolate high-yield, pure RNA from a variety of cell and tissue types, the Direct-zol RNA kit is the optimal solution for RNA extraction from samples in TRIzol®. If you’d like to experience for yourself the power of rapid, scalable RNA extraction, visit our website to learn more about Direct-zol RNA kits and to request your free sample.


