Giving a Whole New Meaning to "Cell" Phone
There is no item more universal and personal than the cell phone. They are a portal to the outside world, with seemingly endless utility. We are constantly poking at them, talking at them, and holding them, so much that they have essentially become an extension of our body. However, that metaphor might be literal when it comes to a cell phone’s microbiome.
The scientists at Zymo Research were curious to find out what microbes might be living on phones, and how geography might influence the abundance of certain types. At Zymo Research’s Global Distributor Meeting, each attendee swabbed their cell phone to help compare these microbiomes from around the globe.
Methods
The samples were collected and processed using the entire ZymoBIOMICS™ workflow from sample collection to conclusion. Swab samples were collected from the cell phones of people from various countries, using the DNA/RNA Shield™ Collection Tube with Swab (Zymo Research, R1106). Tubes were shipped at room temperature before long term storage at -80°C. After storage, the DNA was extracted from each sample using the ZymoBIOMICS™ DNA Microprep Kit (D4301) according to the protocol.
After DNA extraction, sequencing libraries were generated by targeting the V3-4 region of the bacterial 16S rRNA ribosomal RNA gene using the Quick-16S™ NGS Library Prep Kit (D6400) and its V3-4-specific primer set. The amplicon libraries were then quantified and normalized together. Finally, sequencing of the pooled library was performed with a 15% PhiX control library on the Illumina® MiSeq® with the MiSeq® Reagent Kit v3 (600 cycle).
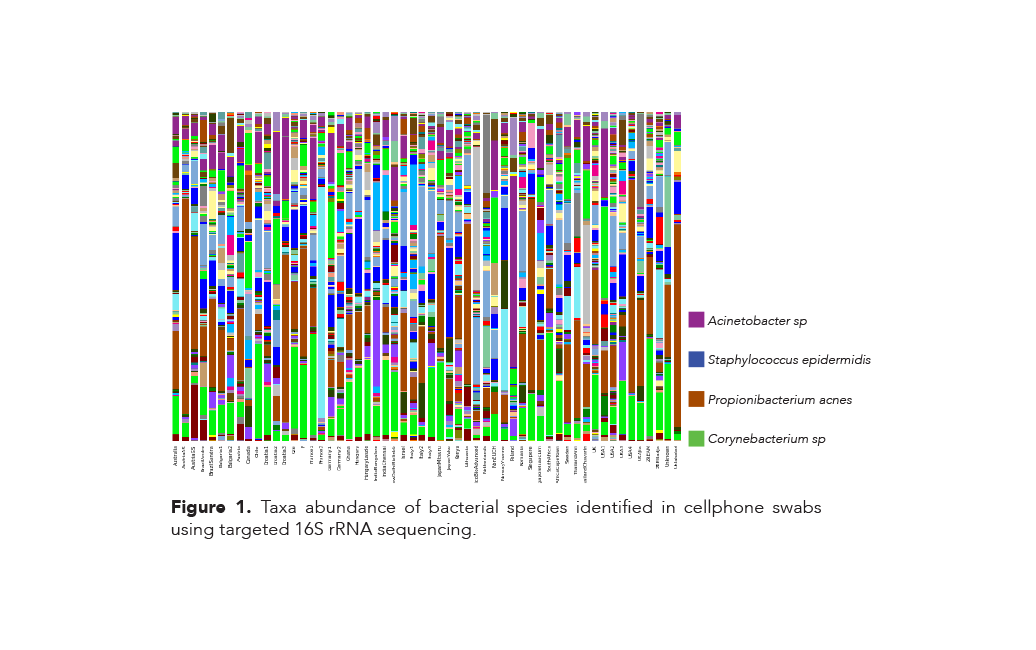
Results
The bacterial species identified and their relative abundances varied widely among the 56 samples that were collected from cell phones of people who lived all over the world (Figure 1). However, three species of bacteria were identified in every cell phone sample tested: Propionibacterium acnes, Staphylococcus epidermis, and a single species of the genus Acinetobacter.
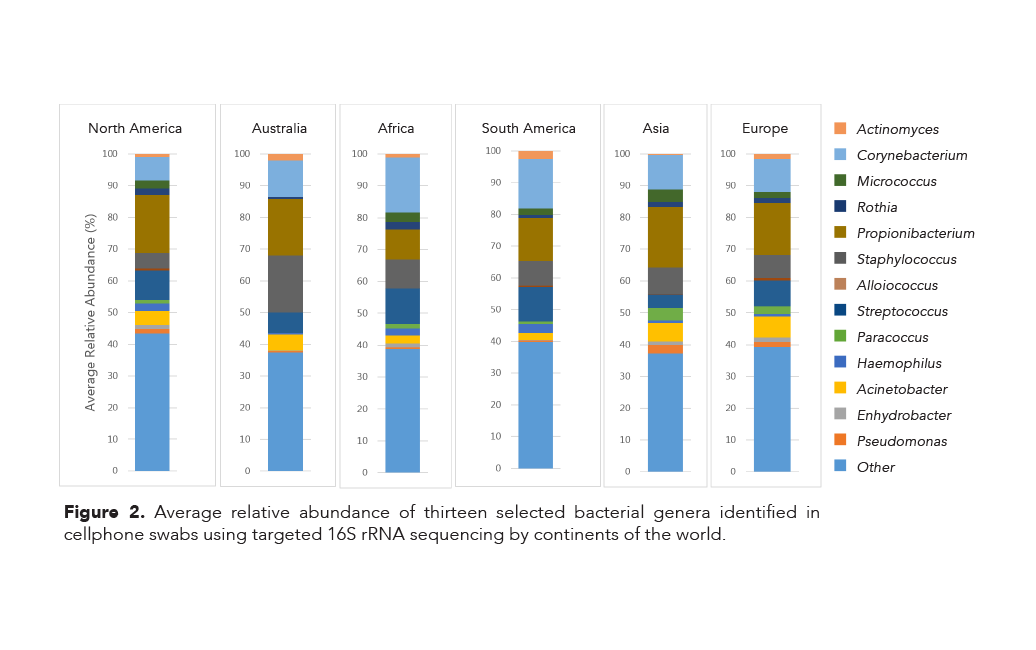
In addition, a single species of the genus Corynebacterium was found in every sample, except for a single swab originating in Canada. All four of these species are associated with normal human skin microflora. Furthermore, thirteen different genera were identified (Figure 2) and while the genera are similar across the continents, there are notable differences in their relative abundances. For example:
- Bacteria in the genus Corynebacterium are identified in similar abundances in samples from Africa and South America; the same trend is seen among samples from Asia and Europe.
- The average relative abundance of bacteria in the genus Propionibacterium is much smaller in samples from Africa than any other continent.
- There is a greater average abundance of bacteria from both Paracoccus and Acinetobacter genera in Asia and Europe than in any other continent.
- Samples from people who resided in North America had the highest relative abundance of bacteria from the genus Pseudomonas
In addition to the similarities and differences found in relative abundances, the identification of several specific microbes led to some interesting findings. For instance, the only isolated species from the genus Alloiococcus, was first isolated from human middle ear fluid and was originally thought to be a pathogenic organism.
Today, new research has shown that it may be a commensal organism in the human microflora2. Interestingly, this organism was identified in most samples from the cell phones belonging to people living in countries in North and South America. However, it was only identified in half of the samples from countries in Europe, and was absent in the majority of samples from Australia, Africa, and Asia (Figure 3).
These results indicate that this species is more prevalent in the normal microflora of people living in countries in the Western Hemisphere and they potentially corroborate the findings that it is a commensal species rather than a pathogenic one.
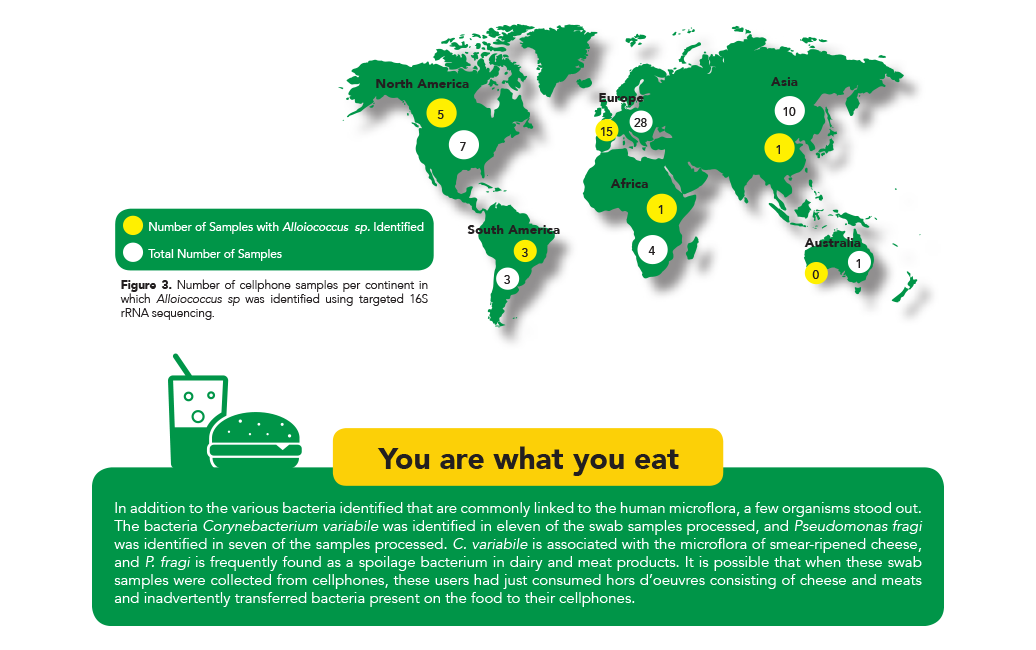
Surprisingly, in addition to the various bacteria identified that are commonly linked to the human microflora, a few organisms stood out. The bacteria Corynebacterium variabile was identified in 11 of the swab samples processed, and Pseudomonas fragi was identified in seven of the samples processed. C. variabile is associated with the microflora of smear-ripened cheese, and P. fragi is frequently found as a spoilage bacterium in dairy and meat products. It is possible that when these swab samples were collected from cell phones, these users had just consumed meat and cheese hors d’oeuvres and inadvertently transferred bacteria present on the food to their cell phones.
Many similarities and differences were discovered among the microbial communities on cell phones from the far corners of the Earth. Because the near-constant use of cell phones has become almost ubiquitous across the globe, some may start to consider the phone’s microbiomes as another component of the human body’s microflora. Going a step further, it is not inconceivable to see the future of microbiome-based medicine include the microflora of the cell phone as an indicator or even a major factor in human health.
References:
1. Davis CP. Normal Flora. In: Baron S, editor. Medical Microbiology. 4th edition. Galveston (TX): University of Texas Medical Branch at Galveston; 1996. Chapter 6. Available from: https://www.ncbi.nlm.nih.gov/books/NBK7617/
2. Tano, Krister et al. “Alloiococcus otitidis- Otitis media pathogen or normal bacterial flora?” Apmis 116 (2008): 785-90. Web. 16 Feb. 2017.


