Getting the Whole Picture
Total Microbial Cell Quantification? Absolutely!
The field of metagenomics has progressed by leaps and bounds over the past decade and even more so with the recent adoption of microbiomics-grade sample collection and unbiased nucleic acid purification techniques. Studies in this field have relied on finding differences in the relative abundance of microbes between samples.
However, understanding relative abundance is only one aspect of the complex nature of microbiomes. For example, what if a treatment doesn’t change the relative abundance, but instead alters the total microbial load? Such a change in the microbiome would go unnoticed with traditional methods. Alternatively, an increase in abundance could be noted for one species, but without additional calculations, it is impossible to infer if the shift is due to an absolute increase in the cell count of that species or a decrease in the cell number of others [1]. Thus, the ability to perform absolute quantification of cell numbers in a sample adds great value to microbiomics and metagenomics. Taking this into consideration, Zymo Research now has their sights set on simplifying absolute cell quantification. The ZymoBIOMICS Spike-in Control enables absolute cell number measurements using Next-Gen sequencing microbial profiling techniques such as 16S rRNA sequencing and shotgun metagenomic sequencing.
A Spike in Interest
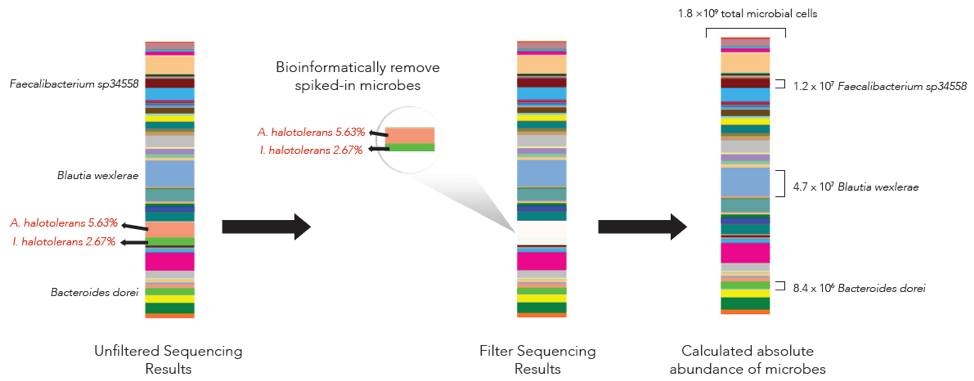
Absolute cell number quantification is achieved by spiking in a known number of microbial cells into a sample, performing NGS-based microbial profiling (Figure 1), and making calculations based on the output of the known cells.
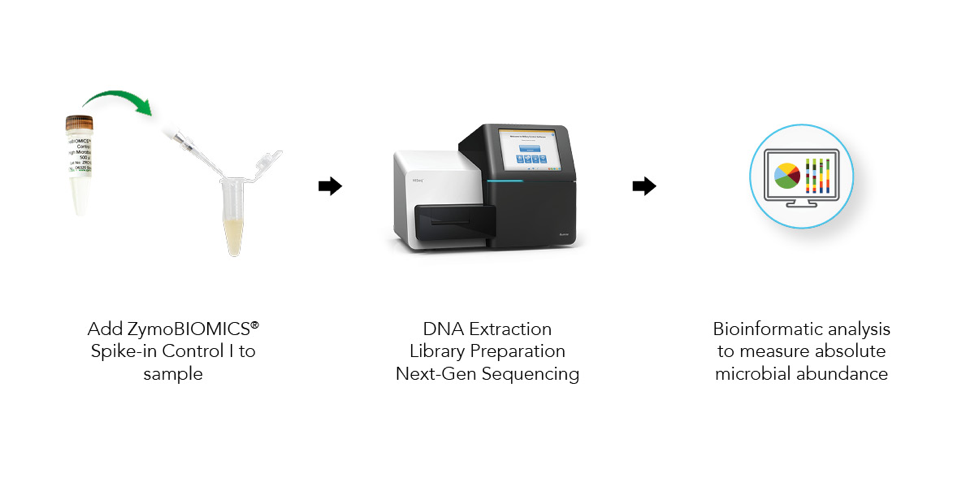
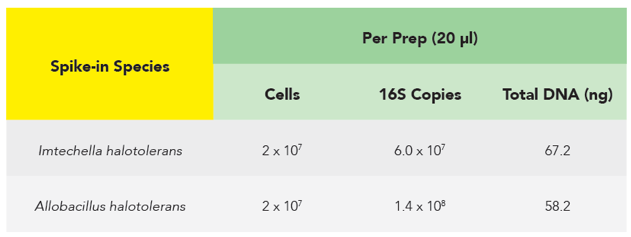
The microbes in the ZymoBIOMICS Spike-in Control are distinguishable from common species because they are only found in very specific and unique environments. Imtechella halotolerans (Gram-negative) and Allobacillus halotolerans (Gram-positive) are tolerant to high salt environments (halotolerant) and are usually only found in brackish water. These exotic species are spiked into samples at known quantities (Table 1) which can then be removed bioinformatically, yielding the original species profile (Figure 2).
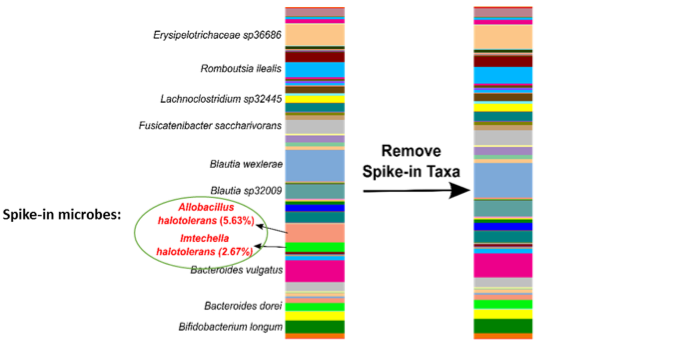
Making Quantification Abundantly Clear
When performing 16S sequencing, calculations are made based on the number of 16S copies per spike-in microbial cells and an average number of 16S copies per unknown cells. For instance, to calculate total cells based on the 16S copies of I. halotolerans, one would use the following formulas:
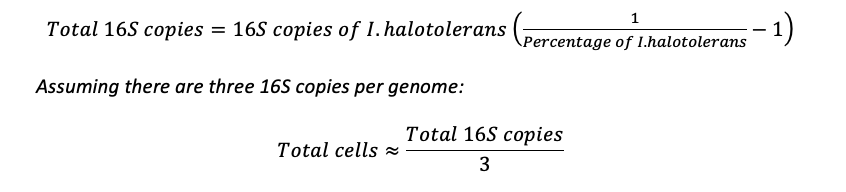
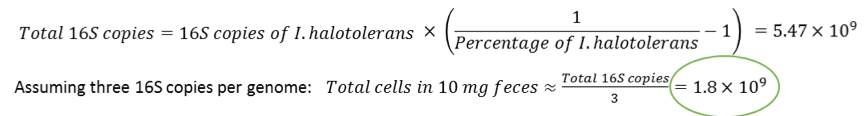
Absolutely what Microbiome Studies Need
The popularity of microbiome profiling has prompted many labs to search for microbial differences among various samples and treatments, but studies are often not published due to the lack of variation between relative abundances. Because of this, standardized solutions are being called for [2, 3] and the field of microbiomics is once again poised to make another leap in progress with the adoption of absolute cell quantification.
This whole-picture microbial profiling is another step in the right direction for metagenomics, illuminating a path toward the most accurate microbial analyses and perhaps one day, personalized microbiome-based medicine. The unique microbial composition of the ZymoBIOMICS™ Spike-in Control allows it to be spiked into almost any microbiome sample, giving researchers the ability to quantify the sample’s total microbial load. This adds a new analytical dimension to microbiome profiling, potentially revealing significant sample differences previously undetected by relative abundance-based analyses.
References:
1. Stämmler F, et al. Adjusting microbiome profiles for differences in microbial load by spike-in bacteria. Microbiome 2016, 4(1):28
2. Jones MB, et al. Library preparation methodology can influence genomic and functional predictions in human microbiome research. PNAS 2015, 112(45) 14024-14029
3. Sinha R, Abnet CC, White O, Knight R, Huttenhower C. The microbiome quality control project: baseline study design and future directions. Genome Biology 2015, 16(276)


