Feces and the Microbiome
The number of human microbiome studies has exploded in the past decade with a majority analyzing fecal samples. While not the most pleasant sample to work with, there is a lot for us to learn about human health and biology that can be found in stool. Contained in these matrices of digested waste and microbes are clues to the many ways that the microbiome affects our health. For example, dysbiosis of the microbial communities we carry in the gut can vary, from obvious infection by foreign pathogens to subtle changes in the commensals that lead to symptoms.
The microbial genomic DNA found in fecal samples is of critical importance to microbiome studies. And while scientists have been continually improving DNA extraction technologies, many challenges in sample purification are still present. These challenges include biased microbial lysis, contamination with PCR inhibitors, and variations in the fecal matrix (hard, soft, diarrhea, etc.). Fecal DNA extractions need to address these major problems to obtain pure, inhibitor-free DNA while maintaining high yields.
Everybody Poops, with Mixed Results
Fecal samples come in all shapes, sizes, and consistencies and can be affected by a variety of factors. The donor species, health, and diet all contribute to the difference in the composition of the fecal material. This results in wide differences in mass, consistency, and microbial community composition.
As such, it is important to have a very robust purification workflow to ensure that microbial DNA can be isolated from all types of samples. The ideal extraction system efficiently lyses both Gram-positive and Gram-negative strains of bacteria as well as archaea and fungi without introducing any bias. On the contrary, using a biased method to isolate bacterial DNA can have drastic consequences for data analysis and lead to incorrect data interpretation. Often, these biases are shown in an inaccurate overrepresentation of the easy-to-lyse organisms, and subsequent underrepresentation of the tough-to-lyse organisms.
Even with the wide variety of issues encountered with many purification workflows, the following tips can ensure successful extraction of DNA from feces:
Breaking It Down
- A little goes a long way
- Each sample has different biomass and bacterial richness, so it is critical to use the appropriate amount of sample for your extraction method. Feces tends to be very rich in bacterial DNA content, so you don’t need much!
- Try starting with as little as 1 mg of healthy stool and work your way up to higher amounts. Typically, 100 mg yields 2.5 ug of DNA
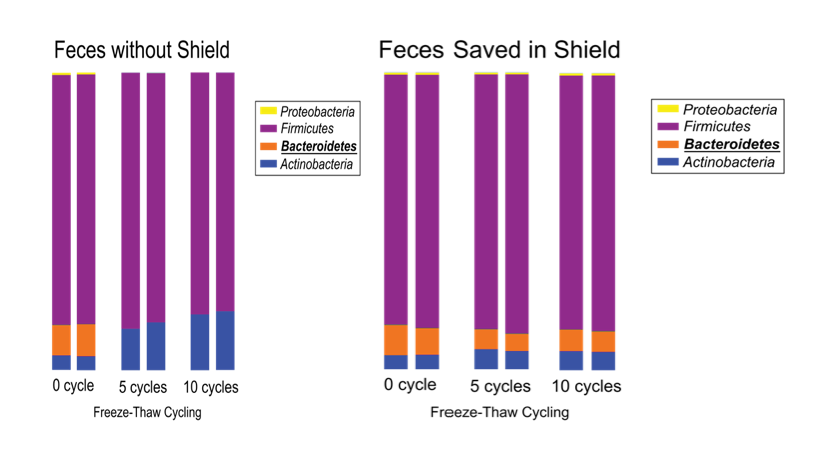
- For very thick or viscous samples, resuspension and dilution into preservation reagents, such as DNA/RNA Shield, will aid in extraction steps while preserving the microbial profile.
- Alternatively, diluting the sample with DNase/RNase free water or a salt solution like PBS also aids extraction. However, the use of water or PBS can lead to microbial cell lysis and release of DNases and RNases and degradation of microbial DNA if extraction is not immediately performed. Additionally, water and PBS will not protect samples from freeze/thaw biases if samples are stored frozen. These samples can also be thoroughly homogenized by passing it through a blender prior to lysis
- Be wary of microbial growth! In the time between collection and transport to a lab for filtration, the microbe populations may change as they grow in your collection tube. There are a few things you can do to ensure growth doesn’t occur and bias your results:
- Use a sample storage reagent (e.g. DNA/RNA Shield)
- Lyse at time of collection
- Freeze sample immediately upon collection (although see caveat above about storage and freeze/thaw bias) (Figure 1)
- For very viscous samples or those higher in inhibitory substances, consider using less sample than usual to avoid having to deal with inhibitors in your PCR.
- Samples from herbivores (e.g. horses and grain fed mice) tend to be very high in carbohydrates and undigested plant matter. Consider treating these samples as you do plants and perform a pre-processing CTAB extraction to remove these carbohydrates before purification.
- Watery stool samples need to be transferred into a lysis tube by pipetting. This is best done with tips that have the ends cut off to make the bore wider for collection of both liquid and particulate matter.
Cleaning Up
DNA purity is critical for successful downstream analyses. If care is not taken during isolation to ensure that the sample is free of contaminants, additional processing steps will be required.
The secret to pure samples lies in the binding and wash steps of any purification protocol. The binding agent contained within most purification kits, including the Quick-DNA Fecal Soil Microbe Kit, is a highly saturated chaotropic salt solution. While it is critical for binding DNA to the purification matrix, this agent will also skew nanodrop measurements by resulting in high absorbances at 230 nm if it co-elutes with the DNA. Additionally, fecal samples can contain high levels of polyphenolics compounds which co-extract the DNA. Care should be taken to ensure that these are removed before steps using PCR as they can inhibit the technique.
To avoid these issues, we recommend the following:
- Do not overfill the column during the binding step of the protocol. Any extra binding buffer can leave salts trapped in the rim or cap of the column. There is also a greater chance of the column tip being contaminated by excess sample loaded.
- During washing steps, the wash buffer should be loaded along the rim of the column rather than simply squirting it into the column. Running the pipette tip along the inner rim of the column helps to wash salts away from the walls of the column.
- Place the elution buffer directly onto the matrix. Pipetting the elution buffer anywhere else on the column may result in resuspension of any residual salts.
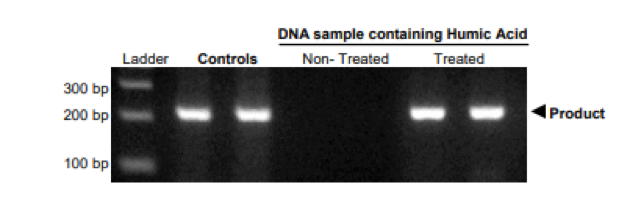
- Fecal samples contain bile salts and complex polysaccharides which copurify with DNA 1, 2. These compounds inhibit PCR and therefore need to be removed for downstream processes (Figure 2). If needed, the eluted DNA should be passed through an inhibitor removal column such as the OneStep PCR Inhibitor Removal Kit.
Citations
- Lantz, P.G. et al. (1997) Removal of PCR inhibitors from human faecal samples through the use of an aqueous two-phase system for sample preparation prior to PCR. J. Microbiol. Meth. 28, 159–67
- Montiero, L. et al. (1997) Complex Polysaccharides as PCR Inhibitors in Feces: Helicobacter pylori Model. J. Clin Microbiol. 35, 995-8
