CTAB Protocol for Isolating DNA From Plant Tissues
The Conventional Plant DNA Extraction Method and a Streamlined Alternative
Modern genomics techniques have promised to revolutionize plant biology, generating data to accelerate crop improvement, optimize plant selection, and advance our basic understanding of plant biology.1 Such techniques and applications rely on the extraction of high-quality DNA from a variety of distinct plant species and sample types.
To keep up with this rapidly advancing field, DNA extraction protocols must be robust, flexible, consistent, and fast. But plant tissues pose several challenges for even the most tried-and-true DNA extraction protocols. Plant cell walls are very difficult to break down and the cells contain many compounds that impede extraction and inhibit downstream molecular biology applications.
The CTAB Method: DNA Extraction from Plant Leaves and Seeds
To overcome the challenges presented by plant tissues, the cetyltrimethylammonium bromide (CTAB) method has become the “go-to” protocol for DNA extraction and purification from leaves and seeds. It was developed in the 1980s and has been used ever since, with various modifications for different plant species.2-5
In the CTAB procedure, the first step is breaking down the tissue, and it involves freezing your plant sample using liquid nitrogen. Once the tissue has been frozen, it’s ground into a fine powder with a mortar and pestle or a blender. After grinding, the tissue is transferred to a tube and CTAB buffer is added. The CTAB buffer facilitates cell lysis and prevents secondary metabolites from interfering with DNA extraction and downstream procedures.
Following plant cell lysis, RNase A is added to digest RNA, and DNA is separated from other cellular components using phenol/chloroform extraction, which separates the sample into two distinct aqueous and organic phases after centrifugation. Nonpolar molecules migrate into the organic phase and leave behind DNA and other polar molecules in the aqueous phase. The extraction is repeated on the aqueous phase until it becomes completely clear, and all DNA is collected. The aqueous phase is collected, and the recovered DNA is precipitated out with isopropanol. The precipitate is pelleted by centrifugation and washed with 70% ethanol to remove salts introduced during extraction. Once the ethanol is decanted, the residual ethanol in the pellet is evaporated away and the dried pellet is resuspended in your buffer-of-choice for your downstream application, such as PCR or NGS.
Considerations Before Performing CTAB DNA Extraction
The CTAB method is biochemically simple, easy to learn, and relatively cheap to perform. However, in practice, the protocol has several drawbacks: it’s lengthy, tedious, and low throughput, with many steps that require careful handling, exposure to hazardous chemicals, and several other technical considerations.
Timing Your DNA Extractions
From grinding with a mortar and pestle to resuspending sticky DNA pellets, the full CTAB protocol can take approximately two hours to process a small number of samples. There are also more than 20 steps in the protocol and as the number of samples increases, the amount of time needed to complete DNA extraction increases substantially. Plan your day carefully and set aside the proper amount of time to complete the entire protocol.
Working With Hazardous Materials
Caution must be taken when working with liquid nitrogen for the first grinding step as it can rapidly freeze skin tissue and cause cold burns even with short exposure. In addition, working with phenol and chloroform is also a biosafety hazard: Phenol can cause chemical burns and chloroform is a potential carcinogen.6,7 For many food testing labs, the use of these toxic chemicals is a major concern. Use of phenol/chloroform also generates organic waste which requires special storage containers and disposal procedures. Be sure you have the proper safety protocols in place before starting your DNA extractions.
Processing the Plant Tissue
Tissue grinding can vary between samples, leading to significant variation in extraction efficiencies and quality of DNA. To achieve more consistent tissue disruption across samples, you can also use a blender, though this step is still low throughput and time consuming. The more finely your tissue is ground, the more efficient you DNA extraction will be, making this a critical step for successful DNA extraction.
Phase Separation
A solution of phenol/chloroform/isoamyl alcohol is used to extract plant DNA from cellular debris and once added and vortexed, the mixture separates into three distinct phases: aqueous, interphase, and organic phase. While removing the aqueous phase and repeating the extraction is time consuming and laborious, it can also be challenging to remove all the aqueous phase, without disturbing the interphase. As a result, you may leave DNA behind or carryover contaminants from the interphase and organic phase, lowering your overall DNA yield and quality.
PCR Inhibitors
Several classes of biochemicals from plant tissues – polysaccharides, lipids, polyphenols, and/or other secondary metabolites – can coprecipitate with DNA, which can inhibit downstream applications that rely on thermostable DNA polymerases, such as PCR. The structure and concentration of these compounds can also vary substantially between different plant species, making the development and optimization of a “one size fits all” CTAB protocol difficult.8,9 In addition, phenol and other salts introduced throughout your CTAB protocol can remain, even after extensive ethanol washes. These impurities can also interfere with downstream applications, including PCR and NGS.10
Zymo’s Alternative Method for Isolation of DNA From Plant Tissues
If that all seems like a bit much, you aren’t wrong. Happily, there are plant-specific DNA isolation kits which provide faster, more consistent, high-purity DNA extraction than the conventional CTAB protocols and variations thereof. These are essential for maintaining and further supporting the rapidly evolving pace, scope, and scale of agricultural R&D.
Zymo’s Quick-DNA Plant/Seed kits use bead beating and column-based purification to provide a simple, rapid workflow for the isolation of inhibitor-free DNA from a variety of plant sources (Figure 1). There are no repetitive and lengthy phase separation steps or hazardous reagents used, so you can further streamline your lab’s operations and protect the safety of key personnel. You can also skip the lengthy RNase digestion, incubation and centrifugation periods, and precipitation steps.
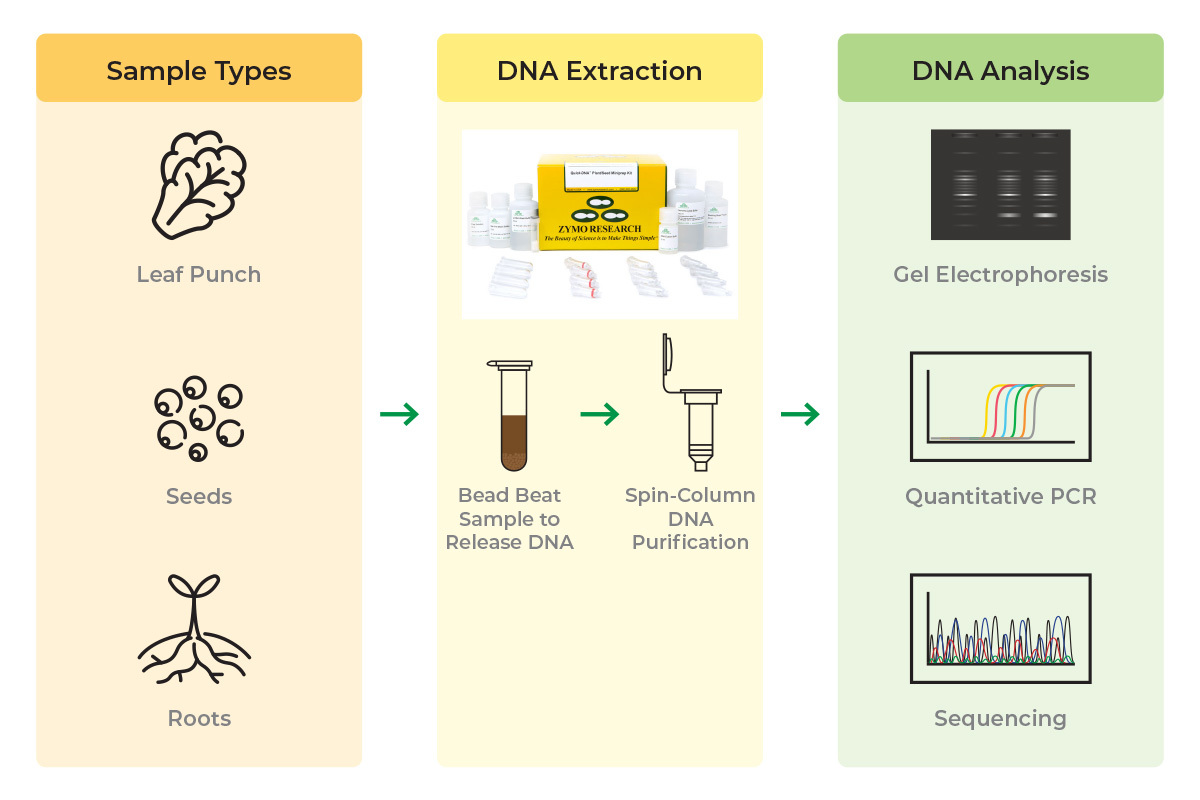
The DNA extraction protocol can be completed in as little as 15 minutes and will give you a straightforward path to high-quality DNA. If you’re working with a challenging plant species, such as cacao and cannabis, try out our state-of-the-art BashingBeads for more complete lysis and more consistent yields (Figure 2).11-14
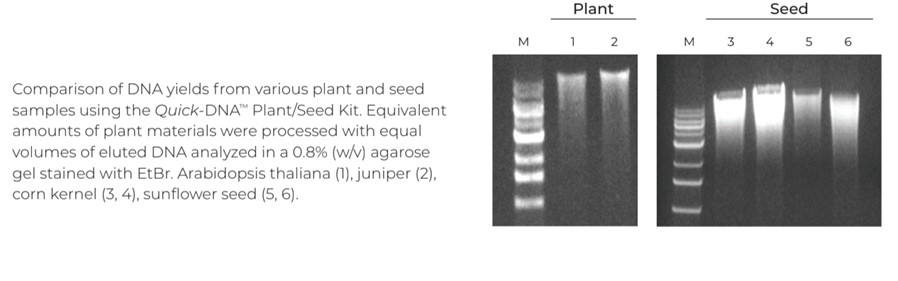
Our binding chemistry, wash solutions, and spin column technology remove polysaccharides, lipids, and other common downstream inhibitors and contaminants providing ultra-pure DNA, with minimal loss. Furthermore, our protocol has been optimized to work with a wide range of plant species and sample types, enabling novel and rapid advancements in modern plant genomics. See how we can help you extract high-quality plant DNA, in less time.
LEARN MORE ABOUT OUR PLANT DNA ISOLATION KITS HERE:
Learn MoreReferences
- Genomics Era for Plants and Crop Species – Advances Made and Needed Tasks Ahead. IntechOpen website: https://www.intechopen.com/chapters/49877. Published July 14th, 2016. Accessed October 27th, 2021.
- Doyle J, Doyle J. A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem Bull. 1987;19(1):11-15.
- Murray MG and Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic acids research. 1980; 8(19); 4321–4325. https://doi.org/10.1093/nar/8.19.4321
- Aboul-Maaty NAF, Oraby HAS. Extraction of high-quality genomic DNA from different plant orders applying a modified CTAB-based method. Bull Natl Res Cent. 2019;43(25). doi.org/10.1186/s42269-019-0066-1
- Muhammad I, Zhang T, Wang Y, et al. Modification of CTAB protocol for maize. Res J Biotech. 2013;8:41–45.
- 22. Safe Use of Phenol | Safety Services. UC - Davis Safety Services website: https://safetyservices.ucdavis.edu/safetynet/safe-use-of-phenol. Published March 26th, 2020. Accessed October 27th, 2021.
- Report on Carcinogens, Fourteenth Edition. National Toxicology Program website: https://ntp.niehs.nih.gov/ntp/roc/content/profiles/chloroform.pdf. Published November 3rd, 2016. Accessed October 27, 2021.
- Aboul-Maaty NAF, Oraby HAS. Extraction of high-quality genomic DNA from different plant orders applying a modified CTAB-based method. Bull Natl Res Cent. 2019;43(25). doi.org/10.1186/s42269-019-0066-1
- CTAB Protocol for the Isolation of DNA from Plant Tissues. OPS Diagnostics website: https://opsdiagnostics.com/notes/protocols/ctab_protocol_for_plants.htm. Accessed October 27, 2021.
- Angeles JGC, Laurena AC, Tecson-Mendoza EM. Extraction of genomic DNA from the lipid-, polysaccharide-, and polyphenol-rich coconut (Cocos nucifera L.). Plant Mol Biol Report. 2012;23(3):297-298. doi:10.1007/BF02772760
- Kamber T, Malpica-López N, Messmer MM, et al. A qPCR Assay for the Fast Detection and Quantification of Colletotrichum lupini. Plants. 2021;10(8):1548. doi:10.3390/PLANTS10081548
- Romero Navarro JA, Phillips-Mora W, Arciniegas-Leal A, et al. Application of Genome Wide Association and Genomic Prediction for Improvement of Cacao Productivity and Resistance to Black and Frosty Pod Diseases. Front Plant Sci. 2017;8:1905. doi:10.3389/fpls.2017.01905
- Cornejo OE, Yee MC, Dominguez V, et al. Population genomic analyses of the chocolate tree, Theobroma cacao L., provide insights into its domestication process. Commun Biol. 2018;1:167. doi:10.1038/s42003-018-0168-6
- Garfinkel AR, Otten M, Crawford S. SNP in Potentially Defunct Tetrahydrocannabinolic Acid Synthase Is a Marker for Cannabigerolic Acid Dominance in Cannabis sativa L. Genes. 2021;12(2):228. doi:10.3390/GENES12020228